You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003162_00531
You are here: Home > Sequence: MGYG000003162_00531
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
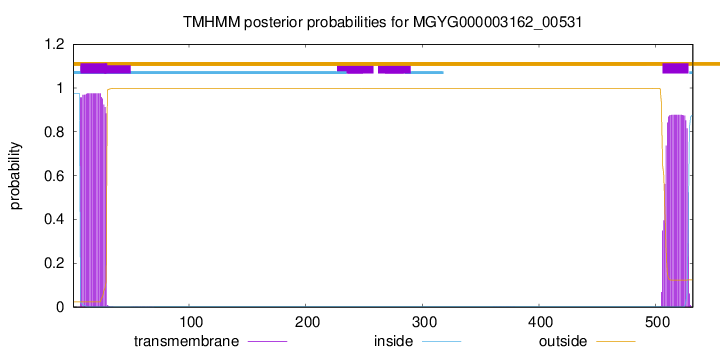
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_D; | |||||||||||
| CAZyme ID | MGYG000003162_00531 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 209; End: 1807 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 90 | 375 | 1.1e-78 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.68e-43 | 89 | 377 | 14 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.39e-12 | 50 | 341 | 46 | 329 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG2723 | BglB | 0.003 | 101 | 163 | 62 | 124 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| NF033846 | Rumino_NPXTG | 0.004 | 505 | 532 | 1 | 29 | NPXTG family C-terminal sorting domain. Rumino_NPXTG represents a flavor of C-terminal protein sorting signal in species related to Ruminococcus albus. In that lineage, multiple sortases per genome may be found, including multiple B-type sortases. Proteins found by this HMM (more than 12 encoded in a representative complete genome) may represent substrates of a panel of related sortases, while additional proteins found in Ruminococcus genomes with below-cutoff hits to this model may be processed by other sortases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL35246.1 | 3.63e-147 | 5 | 402 | 3 | 408 |
| CBK97722.1 | 8.33e-146 | 36 | 402 | 44 | 408 |
| CCO05195.1 | 3.25e-71 | 36 | 394 | 41 | 392 |
| CBL35076.1 | 8.70e-67 | 20 | 388 | 4 | 364 |
| CBK96759.1 | 2.26e-66 | 1 | 388 | 1 | 374 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6WQP_A | 1.26e-52 | 41 | 375 | 9 | 324 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 6Q1I_A | 1.90e-52 | 47 | 383 | 13 | 325 | GH5-4broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum],6Q1I_B GH5-4 broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum] |
| 6WQY_A | 1.96e-52 | 47 | 404 | 23 | 377 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
| 6XRK_A | 1.15e-47 | 47 | 381 | 23 | 353 | GH5-4broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate],6XRK_B GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate] |
| 5D9N_A | 3.63e-47 | 42 | 372 | 2 | 324 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9N_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9P_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii],5D9P_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54937 | 4.99e-51 | 47 | 383 | 38 | 350 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P17901 | 7.46e-46 | 57 | 408 | 56 | 409 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| O08342 | 1.11e-43 | 46 | 377 | 38 | 371 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P16216 | 8.88e-43 | 30 | 393 | 51 | 384 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
| P23660 | 3.42e-41 | 40 | 383 | 20 | 337 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
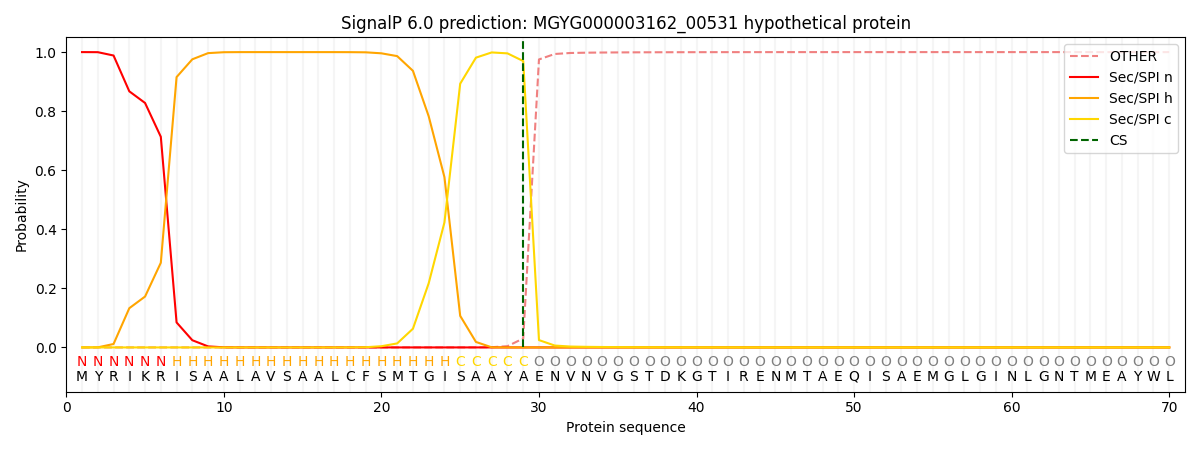
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000509 | 0.998513 | 0.000238 | 0.000280 | 0.000234 | 0.000211 |