You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003099_01109
You are here: Home > Sequence: MGYG000003099_01109
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Barnesiella sp900538555 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Barnesiellaceae; Barnesiella; Barnesiella sp900538555 | |||||||||||
| CAZyme ID | MGYG000003099_01109 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 69790; End: 72408 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 70 | 313 | 1.9e-68 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 2.58e-124 | 63 | 861 | 92 | 756 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 2.48e-118 | 9 | 828 | 6 | 726 | Probable beta-xylosidase; Provisional |
| COG1472 | BglX | 3.06e-70 | 38 | 411 | 14 | 366 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 4.85e-56 | 384 | 754 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam00933 | Glyco_hydro_3 | 1.29e-46 | 72 | 344 | 63 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT77714.1 | 0.0 | 11 | 872 | 6 | 863 |
| QDO68996.1 | 0.0 | 16 | 872 | 5 | 864 |
| QUT88122.1 | 0.0 | 16 | 872 | 6 | 863 |
| ALJ60860.1 | 0.0 | 16 | 872 | 6 | 863 |
| QUT40961.1 | 0.0 | 16 | 872 | 6 | 863 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 1.00e-90 | 60 | 869 | 108 | 784 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 7VC7_A | 2.10e-89 | 43 | 815 | 22 | 677 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC6_A | 2.10e-89 | 43 | 815 | 22 | 677 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 6R5I_A | 1.44e-80 | 71 | 869 | 68 | 732 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5O_A | 7.30e-80 | 71 | 869 | 68 | 732 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5O_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5R_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 2.01e-305 | 27 | 871 | 15 | 861 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| Q9SGZ5 | 1.78e-100 | 45 | 858 | 47 | 759 | Probable beta-D-xylosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BXL7 PE=2 SV=2 |
| Q0CB82 | 1.79e-97 | 9 | 853 | 8 | 730 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=bxlB PE=3 SV=1 |
| A1DJS5 | 1.50e-96 | 13 | 853 | 18 | 738 | Probable exo-1,4-beta-xylosidase xlnD OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xlnD PE=3 SV=1 |
| B0Y0I4 | 2.92e-95 | 38 | 872 | 44 | 757 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=bxlB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
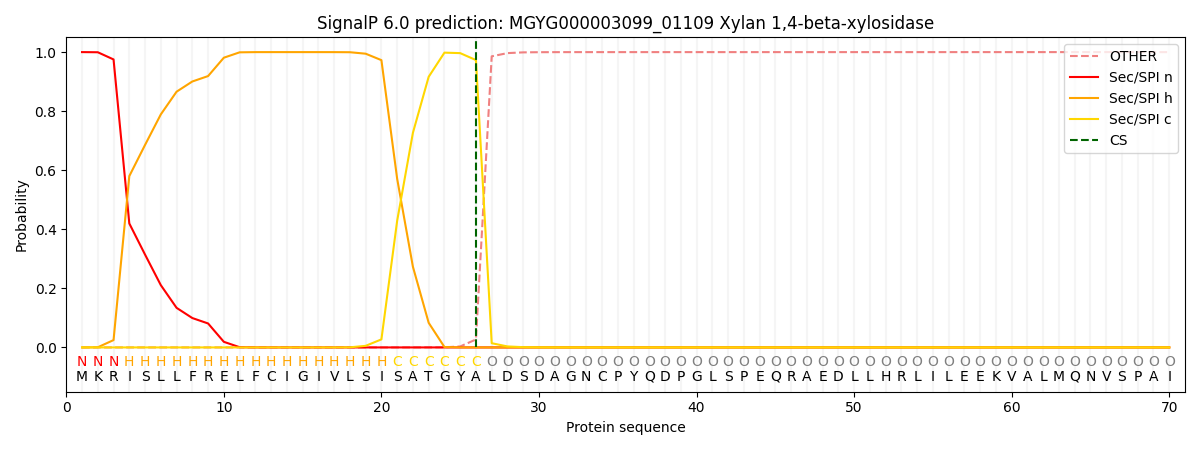
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000274 | 0.999039 | 0.000192 | 0.000159 | 0.000153 | 0.000145 |
