You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003072_02668
You are here: Home > Sequence: MGYG000003072_02668
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
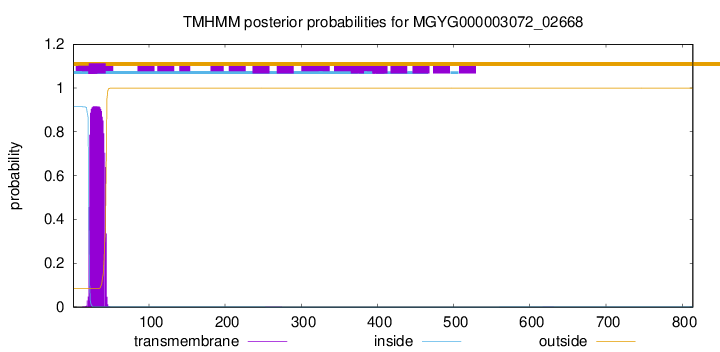
TMHMM annotations
Basic Information help
| Species | Paenibacillus amylolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus amylolyticus | |||||||||||
| CAZyme ID | MGYG000003072_02668 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 227036; End: 229480 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 73 | 418 | 5.5e-98 | 0.9421768707482994 |
| CBM3 | 678 | 744 | 3.9e-18 | 0.8181818181818182 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5297 | CelA1 | 0.0 | 23 | 473 | 1 | 446 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| pfam01341 | Glyco_hydro_6 | 1.99e-135 | 76 | 420 | 1 | 292 | Glycosyl hydrolases family 6. |
| pfam00942 | CBM_3 | 6.71e-18 | 677 | 749 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 8.55e-16 | 677 | 750 | 1 | 83 | Cellulose binding domain. |
| COG3401 | FN3 | 1.55e-07 | 501 | 663 | 181 | 341 | Fibronectin type 3 domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZN78074.1 | 0.0 | 1 | 814 | 1 | 814 |
| APO48137.1 | 0.0 | 15 | 814 | 1 | 800 |
| QLG42501.1 | 0.0 | 3 | 814 | 2 | 810 |
| QKS58655.1 | 0.0 | 1 | 814 | 1 | 814 |
| AQM58155.1 | 0.0 | 1 | 814 | 1 | 814 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XYH_A | 2.41e-224 | 45 | 470 | 1 | 425 | CrystalStructure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae [Xanthomonas oryzae pv. oryzae] |
| 7CBD_A | 6.85e-174 | 44 | 470 | 2 | 431 | Catalyticdomain of Cellulomonas fimi Cel6B [Cellulomonas fimi ATCC 484] |
| 6K55_A | 4.72e-151 | 48 | 470 | 2 | 426 | Inactivatedmutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide [Ardenticatenia bacterium],6K55_B Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide [Ardenticatenia bacterium],6K55_C Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide [Ardenticatenia bacterium] |
| 4B4F_A | 3.18e-126 | 48 | 470 | 3 | 420 | Thermobifidafusca Cel6B(E3) co-crystallized with cellobiose [Thermobifida fusca],4B4F_B Thermobifida fusca Cel6B(E3) co-crystallized with cellobiose [Thermobifida fusca],4B4H_A Thermobifida fusca cellobiohydrolase Cel6B(E3) catalytic domain [Thermobifida fusca],4B4H_B Thermobifida fusca cellobiohydrolase Cel6B(E3) catalytic domain [Thermobifida fusca] |
| 4AVO_A | 4.93e-125 | 48 | 470 | 3 | 420 | Thermobifidafusca cellobiohydrolase Cel6B catalytic mutant D274A cocrystallized with cellobiose [Thermobifida fusca YX] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P50401 | 5.92e-180 | 41 | 611 | 38 | 616 | Exoglucanase A OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhA PE=1 SV=1 |
| P49075 | 8.40e-57 | 50 | 470 | 90 | 438 | Exoglucanase 3 OS=Agaricus bisporus OX=5341 GN=cel3 PE=1 SV=1 |
| P29718 | 1.45e-56 | 677 | 814 | 1 | 145 | Uncharacterized protein in celA 5'region (Fragment) OS=Paenibacillus lautus OX=1401 PE=4 SV=1 |
| P07987 | 6.07e-55 | 50 | 468 | 115 | 468 | Exoglucanase 2 OS=Hypocrea jecorina OX=51453 GN=cbh2 PE=1 SV=1 |
| A0A024SH76 | 6.07e-55 | 50 | 468 | 115 | 468 | Exoglucanase 2 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=cbh2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
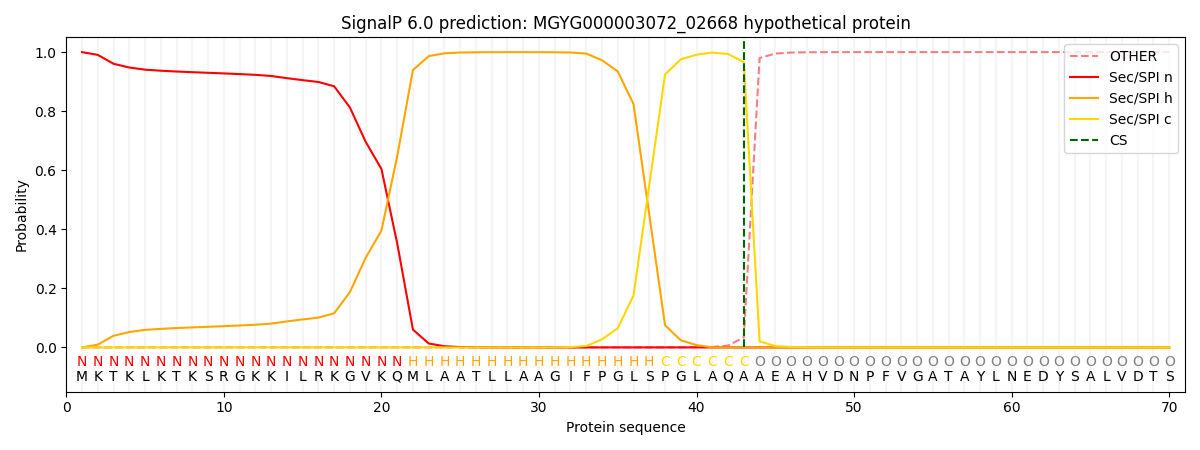
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000395 | 0.998771 | 0.000248 | 0.000228 | 0.000176 | 0.000147 |