You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003072_00048
You are here: Home > Sequence: MGYG000003072_00048
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
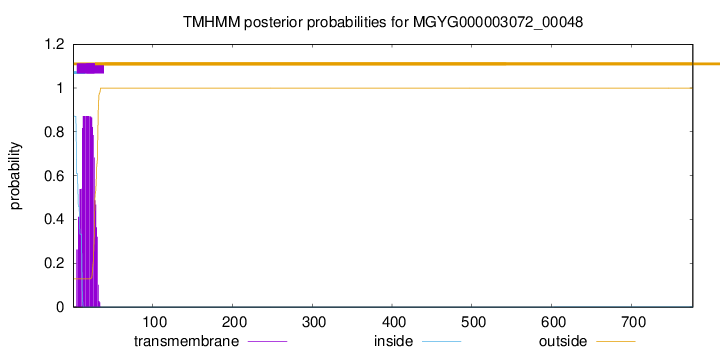
TMHMM annotations
Basic Information help
| Species | Paenibacillus amylolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus amylolyticus | |||||||||||
| CAZyme ID | MGYG000003072_00048 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48221; End: 50554 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 199 | 521 | 1.9e-116 | 0.9933993399339934 |
| CBM22 | 38 | 168 | 1.4e-33 | 0.9847328244274809 |
| CBM3 | 631 | 710 | 7.4e-24 | 0.9659090909090909 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 2.37e-134 | 200 | 521 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.02e-127 | 241 | 519 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.29e-102 | 181 | 521 | 5 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00942 | CBM_3 | 1.51e-23 | 631 | 710 | 3 | 82 | Cellulose binding domain. |
| pfam02018 | CBM_4_9 | 4.95e-23 | 38 | 169 | 3 | 132 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APO45385.1 | 0.0 | 1 | 777 | 1 | 777 |
| AWV32895.1 | 0.0 | 1 | 775 | 1 | 770 |
| ANY65885.1 | 0.0 | 6 | 775 | 9 | 781 |
| QRK10473.1 | 0.0 | 32 | 775 | 28 | 672 |
| ATB37912.1 | 4.28e-316 | 19 | 775 | 18 | 674 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NL2_A | 8.15e-111 | 189 | 523 | 1 | 341 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 6D5C_A | 1.66e-105 | 197 | 521 | 20 | 349 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 5OFJ_A | 8.93e-105 | 183 | 521 | 1 | 337 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
| 5OFK_A | 6.90e-104 | 183 | 521 | 1 | 337 | Crystalstructure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose [Caldicellulosiruptor bescii DSM 6725],5OFL_A Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose [Caldicellulosiruptor bescii DSM 6725] |
| 6FHE_A | 1.24e-100 | 194 | 520 | 7 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10474 | 1.59e-137 | 197 | 775 | 42 | 570 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| Q60037 | 2.14e-115 | 38 | 533 | 205 | 700 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| Q60042 | 1.06e-113 | 38 | 533 | 200 | 696 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| P36917 | 9.06e-98 | 38 | 521 | 197 | 674 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| O69230 | 1.69e-97 | 44 | 521 | 205 | 709 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
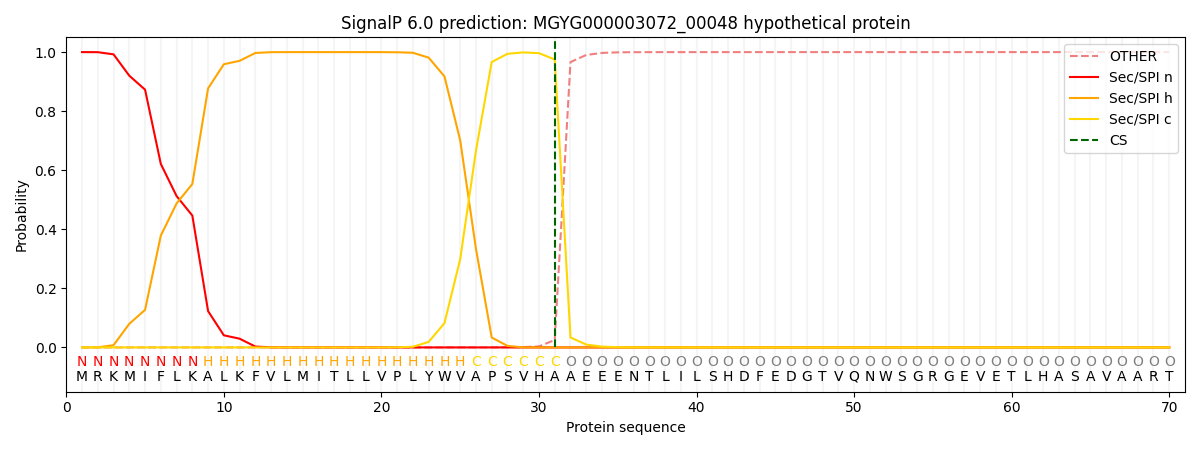
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000266 | 0.999074 | 0.000190 | 0.000163 | 0.000146 | 0.000134 |