You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003064_01867
You are here: Home > Sequence: MGYG000003064_01867
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides graminisolvens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides graminisolvens | |||||||||||
| CAZyme ID | MGYG000003064_01867 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 176785; End: 179004 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 493 | 722 | 2.9e-53 | 0.6600660066006601 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.18e-46 | 493 | 722 | 102 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 4.74e-46 | 482 | 720 | 50 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 9.31e-34 | 493 | 727 | 125 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 6.68e-10 | 55 | 147 | 1 | 92 | Glycosyl hydrolase family 10. |
| pfam02018 | CBM_4_9 | 2.63e-07 | 165 | 280 | 2 | 115 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDH57041.1 | 9.66e-237 | 1 | 738 | 1 | 740 |
| ALJ46369.1 | 3.13e-235 | 1 | 738 | 1 | 740 |
| QRQ57932.1 | 3.13e-235 | 1 | 738 | 1 | 740 |
| QGT71452.1 | 4.44e-235 | 1 | 738 | 1 | 740 |
| QUT80661.1 | 3.58e-234 | 1 | 738 | 1 | 740 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4W8L_A | 1.29e-26 | 495 | 722 | 113 | 343 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 4XUY_A | 2.32e-26 | 495 | 721 | 109 | 299 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
| 3MMD_A | 6.61e-26 | 481 | 710 | 122 | 336 | ChainA, Endo-1,4-beta-xylanase [Geobacillus stearothermophilus] |
| 6FHE_A | 1.14e-25 | 495 | 717 | 130 | 335 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 1HIZ_A | 1.62e-25 | 481 | 710 | 122 | 336 | XylanaseT6 (Xt6) from Bacillus Stearothermophilus [Geobacillus stearothermophilus],1R85_A Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6): The WT enzyme (monoclinic form) at 1.45A resolution [Geobacillus stearothermophilus],1R87_A Crystal structure of the extracellular xylanase from Geobacillus stearothermophilus T-6 (XT6, monoclinic form): The complex of the WT enzyme with xylopentaose at 1.67A resolution [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W0HFK8 | 1.98e-26 | 493 | 721 | 136 | 328 | Endo-1,4-beta-xylanase 1 OS=Rhizopus oryzae OX=64495 GN=xyn1 PE=1 SV=1 |
| P33559 | 6.14e-26 | 495 | 721 | 134 | 324 | Endo-1,4-beta-xylanase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xynA PE=1 SV=2 |
| P36917 | 6.32e-26 | 147 | 722 | 23 | 674 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| C5J411 | 1.12e-25 | 495 | 721 | 134 | 324 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
| Q6PRW6 | 1.77e-25 | 493 | 717 | 136 | 324 | Endo-1,4-beta-xylanase OS=Penicillium chrysogenum OX=5076 GN=Xyn PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
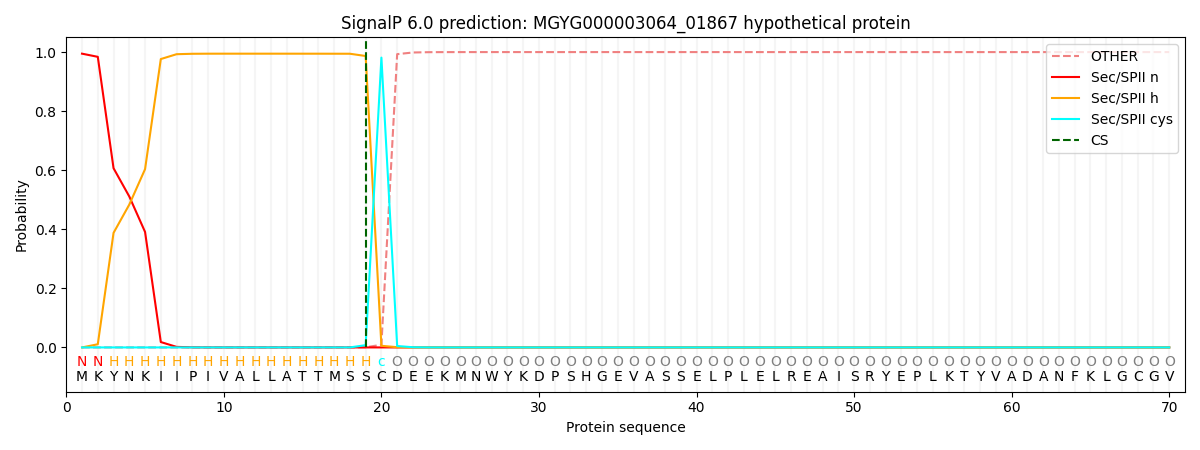
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000008 | 0.005263 | 0.994782 | 0.000002 | 0.000003 | 0.000002 |
