You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003042_01503
You are here: Home > Sequence: MGYG000003042_01503
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
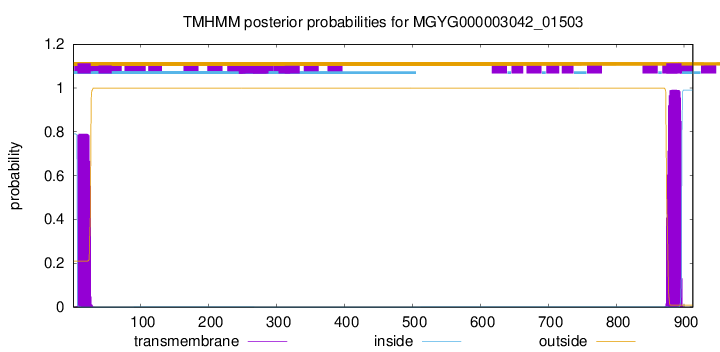
TMHMM annotations
Basic Information help
| Species | Gemmiger sp900545545 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Gemmiger; Gemmiger sp900545545 | |||||||||||
| CAZyme ID | MGYG000003042_01503 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 90870; End: 93611 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 142 | 315 | 1.4e-16 | 0.6898148148148148 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01915 | Glyco_hydro_3_C | 3.74e-17 | 670 | 831 | 60 | 212 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK15098 | PRK15098 | 8.81e-10 | 133 | 487 | 105 | 422 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 4.00e-06 | 706 | 808 | 488 | 589 | Probable beta-xylosidase; Provisional |
| pfam00933 | Glyco_hydro_3 | 2.60e-05 | 143 | 235 | 83 | 162 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYA77556.1 | 1.12e-149 | 46 | 850 | 41 | 843 |
| QKS56187.1 | 9.08e-143 | 33 | 850 | 29 | 841 |
| QMV43973.1 | 5.02e-127 | 38 | 851 | 6 | 818 |
| AYQ75660.1 | 4.19e-116 | 44 | 849 | 38 | 831 |
| BBF43857.1 | 1.42e-90 | 48 | 850 | 41 | 816 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6R5I_A | 1.47e-11 | 133 | 485 | 73 | 388 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 7VC7_A | 2.57e-11 | 129 | 487 | 78 | 399 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC6_A | 2.57e-11 | 129 | 487 | 78 | 399 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 6R5O_A | 5.79e-11 | 133 | 485 | 73 | 388 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5O_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5R_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5R_A | 5.80e-11 | 133 | 485 | 74 | 389 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SGZ5 | 1.43e-10 | 705 | 808 | 481 | 583 | Probable beta-D-xylosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BXL7 PE=2 SV=2 |
| Q9LJN4 | 1.90e-10 | 705 | 808 | 481 | 583 | Probable beta-D-xylosidase 5 OS=Arabidopsis thaliana OX=3702 GN=BXL5 PE=2 SV=2 |
| Q56078 | 4.27e-10 | 133 | 527 | 105 | 461 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
| A5JTQ2 | 2.92e-09 | 699 | 808 | 485 | 593 | Beta-xylosidase/alpha-L-arabinofuranosidase 1 (Fragment) OS=Medicago sativa subsp. varia OX=36902 GN=Xyl1 PE=1 SV=1 |
| P83344 | 4.42e-09 | 706 | 821 | 169 | 285 | Putative beta-D-xylosidase (Fragment) OS=Prunus persica OX=3760 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
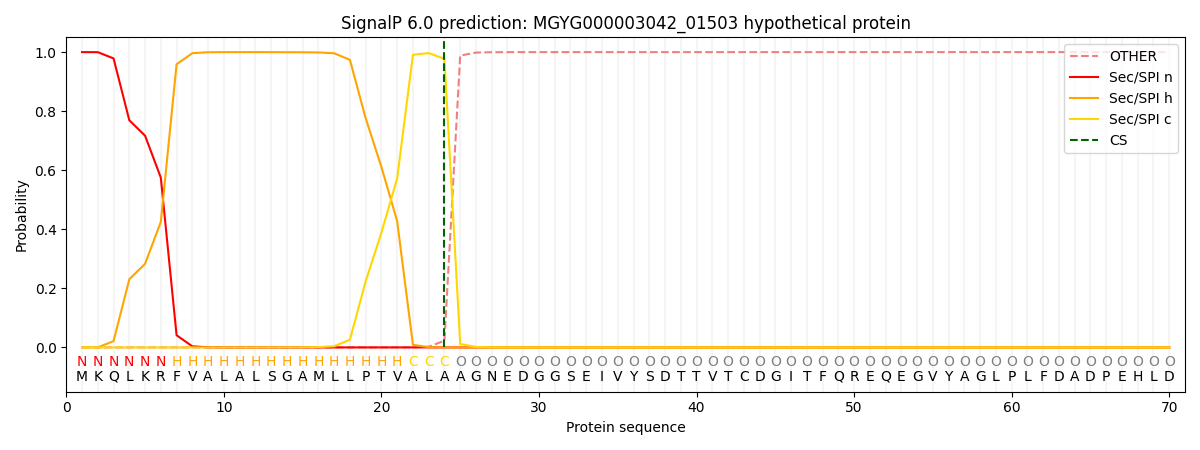
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000270 | 0.999020 | 0.000194 | 0.000185 | 0.000172 | 0.000151 |