You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003035_01267
You are here: Home > Sequence: MGYG000003035_01267
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900542185 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900542185 | |||||||||||
| CAZyme ID | MGYG000003035_01267 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42939; End: 44087 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 43 | 372 | 3.1e-52 | 0.976897689768977 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 5.39e-45 | 48 | 372 | 11 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.44e-36 | 51 | 378 | 39 | 345 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| smart00633 | Glyco_10 | 1.89e-35 | 80 | 370 | 2 | 263 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT43884.1 | 5.52e-149 | 36 | 376 | 52 | 394 |
| QUT22690.1 | 5.14e-134 | 34 | 374 | 37 | 380 |
| QDU59060.1 | 4.25e-103 | 31 | 374 | 52 | 387 |
| QHI69082.1 | 9.87e-103 | 31 | 374 | 22 | 357 |
| QZT37897.1 | 1.94e-97 | 14 | 374 | 32 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VBR_A | 2.56e-24 | 46 | 376 | 14 | 322 | Crystalstructure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBR_B Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBU_A Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima],1VBU_B Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima] |
| 3NIY_A | 3.11e-24 | 46 | 376 | 30 | 338 | Crystalstructure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
| 5AY7_A | 3.75e-19 | 39 | 371 | 19 | 344 | Apsychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5AY7_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5D4Y_A A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples],5D4Y_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples] |
| 5Y3X_A | 1.30e-18 | 35 | 370 | 29 | 354 | Crystalstructure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_B Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_C Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_D Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_E Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL],5Y3X_F Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis [Caldicellulosiruptor owensensis OL] |
| 1E0W_A | 1.97e-16 | 48 | 374 | 15 | 300 | Xylanase10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution [Streptomyces lividans],1E0X_A Xylanase 10a From Sreptomyces Lividans. Xylobiosyl-Enzyme Intermediate At 1.65 A [Streptomyces lividans],1E0X_B Xylanase 10a From Sreptomyces Lividans. Xylobiosyl-Enzyme Intermediate At 1.65 A [Streptomyces lividans],1OD8_A Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine lactam [Streptomyces lividans],1V0K_A Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 5.8 [Streptomyces lividans],1V0L_A Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 [Streptomyces lividans],1V0M_A Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 [Streptomyces lividans],1V0N_A Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 [Streptomyces lividans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q60041 | 8.00e-22 | 29 | 381 | 19 | 346 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
| P40944 | 4.42e-21 | 35 | 370 | 353 | 675 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor sp. (strain Rt8B.4) OX=28238 GN=xynA PE=3 SV=1 |
| P26514 | 3.85e-16 | 2 | 374 | 10 | 341 | Endo-1,4-beta-xylanase A OS=Streptomyces lividans OX=1916 GN=xlnA PE=1 SV=2 |
| P23556 | 7.91e-16 | 41 | 349 | 19 | 311 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynA PE=1 SV=1 |
| Q02290 | 1.08e-15 | 41 | 378 | 25 | 329 | Endo-1,4-beta-xylanase B OS=Neocallimastix patriciarum OX=4758 GN=xynB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
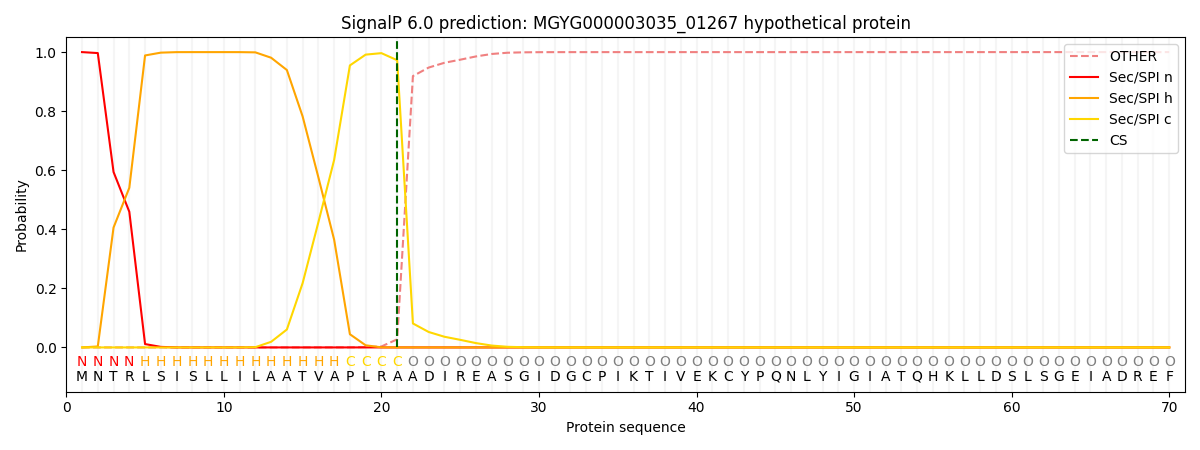
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000231 | 0.999137 | 0.000163 | 0.000152 | 0.000145 | 0.000131 |
