You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002960_00903
You are here: Home > Sequence: MGYG000002960_00903
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900555035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900555035 | |||||||||||
| CAZyme ID | MGYG000002960_00903 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18093; End: 20321 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 464 | 727 | 1.2e-43 | 0.6798679867986799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.18e-26 | 466 | 727 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.36e-24 | 463 | 729 | 93 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.51e-16 | 463 | 724 | 116 | 328 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.37e-09 | 56 | 141 | 5 | 85 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 5.35e-05 | 91 | 144 | 58 | 111 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ61518.1 | 1.75e-261 | 10 | 731 | 8 | 718 |
| QNT67590.1 | 2.55e-259 | 1 | 742 | 1 | 734 |
| AVM57878.1 | 3.72e-242 | 10 | 731 | 5 | 746 |
| AVM52702.1 | 3.72e-242 | 10 | 731 | 5 | 746 |
| QDO69412.1 | 2.52e-203 | 3 | 731 | 8 | 739 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4W8L_A | 2.21e-11 | 458 | 698 | 101 | 308 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 1W2V_A | 1.61e-10 | 478 | 698 | 109 | 310 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W2V_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1CLX_A | 2.83e-10 | 478 | 698 | 108 | 309 | CatalyticCore Of Xylanase A [Cellvibrio japonicus],1CLX_B Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_C Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_D Catalytic Core Of Xylanase A [Cellvibrio japonicus] |
| 1W2P_A | 2.85e-10 | 478 | 698 | 109 | 310 | The3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W2P_B The 3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W32_A | 2.85e-10 | 478 | 698 | 109 | 310 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W32_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O69230 | 3.64e-10 | 458 | 698 | 467 | 674 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| P14768 | 3.25e-09 | 478 | 698 | 372 | 573 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
| P07986 | 1.71e-07 | 472 | 733 | 143 | 355 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| C6CRV0 | 2.08e-07 | 457 | 741 | 615 | 859 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P36917 | 4.48e-07 | 467 | 645 | 459 | 603 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
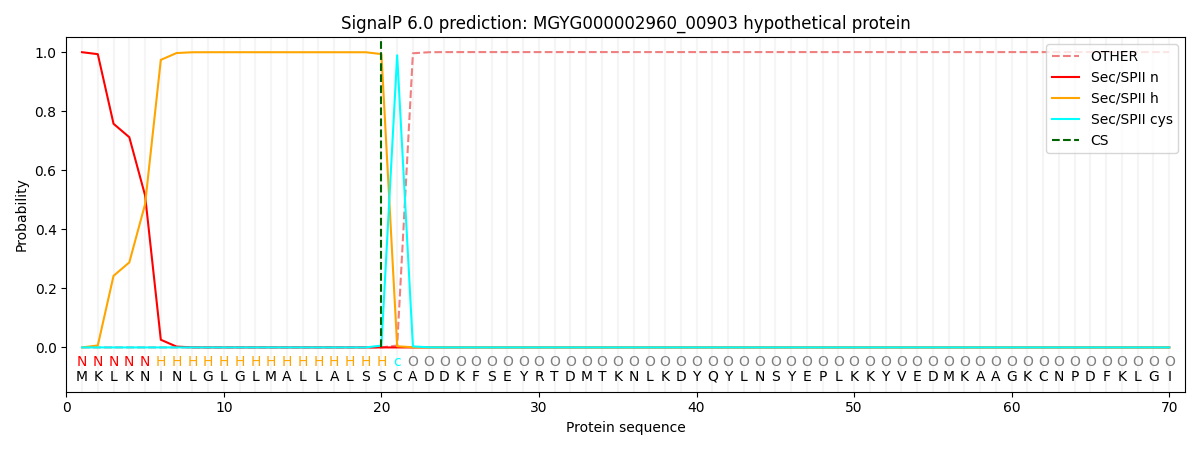
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000425 | 0.999613 | 0.000000 | 0.000000 | 0.000000 |
