You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002943_00517
You are here: Home > Sequence: MGYG000002943_00517
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
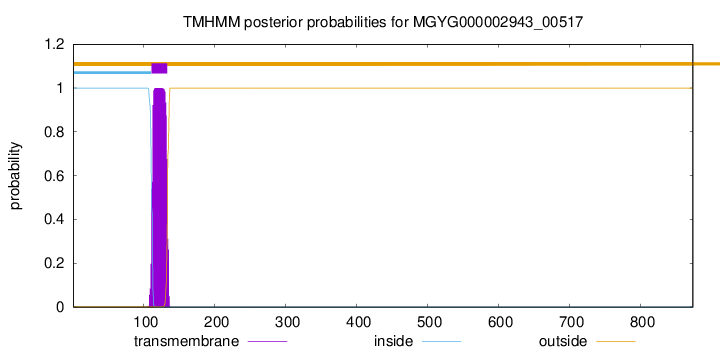
TMHMM annotations
Basic Information help
| Species | Actinomyces oris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces; Actinomyces oris | |||||||||||
| CAZyme ID | MGYG000002943_00517 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3665; End: 6289 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 159 | 331 | 5.4e-62 | 0.9774011299435028 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 1.93e-154 | 97 | 764 | 3 | 650 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 5.30e-114 | 114 | 792 | 11 | 795 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 2.43e-79 | 159 | 332 | 3 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG4953 | PbpC | 3.48e-57 | 139 | 679 | 26 | 531 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| PRK11636 | mrcA | 1.84e-55 | 152 | 730 | 50 | 795 | penicillin-binding protein 1a; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQQ59301.1 | 0.0 | 32 | 848 | 1 | 817 |
| QQC39442.1 | 0.0 | 32 | 805 | 1 | 774 |
| QLF52334.1 | 0.0 | 32 | 808 | 1 | 777 |
| BAV84047.1 | 0.0 | 32 | 763 | 1 | 734 |
| AMD98829.1 | 0.0 | 32 | 745 | 1 | 716 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZG8_B | 2.85e-46 | 251 | 687 | 2 | 436 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 4OON_A | 1.84e-44 | 152 | 719 | 23 | 722 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3ZG7_B | 3.64e-44 | 251 | 687 | 2 | 436 | CrystalStructure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the apo form [Listeria monocytogenes] |
| 3DWK_A | 5.20e-44 | 170 | 681 | 28 | 555 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 5FGZ_A | 7.75e-42 | 169 | 677 | 156 | 662 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0R7G2 | 1.04e-100 | 122 | 744 | 83 | 706 | Penicillin-binding protein 1A OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=ponA1 PE=3 SV=1 |
| P71707 | 8.70e-93 | 122 | 761 | 145 | 785 | Penicillin-binding protein 1A OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=ponA1 PE=1 SV=3 |
| P38050 | 6.61e-63 | 107 | 721 | 10 | 599 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| Q5FAC7 | 7.96e-62 | 151 | 739 | 51 | 725 | Penicillin-binding protein 1A OS=Neisseria gonorrhoeae (strain ATCC 700825 / FA 1090) OX=242231 GN=mrcA PE=3 SV=3 |
| O05131 | 7.96e-62 | 151 | 739 | 51 | 725 | Penicillin-binding protein 1A OS=Neisseria gonorrhoeae OX=485 GN=mrcA PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000065 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |