You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002834_00922
Basic Information
help
| Species |
Prevotella sp900551985
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900551985
|
| CAZyme ID |
MGYG000002834_00922
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002834 |
3427886 |
MAG |
United Republic of Tanzania |
Africa |
|
| Gene Location |
Start: 7587;
End: 9008
Strand: +
|
No EC number prediction in MGYG000002834_00922.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
50 |
320 |
2.9e-106 |
0.9884169884169884 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
2.17e-11 |
149 |
285 |
71 |
203 |
Cellulase (glycosyl hydrolase family 5). |
This protein is predicted as SP
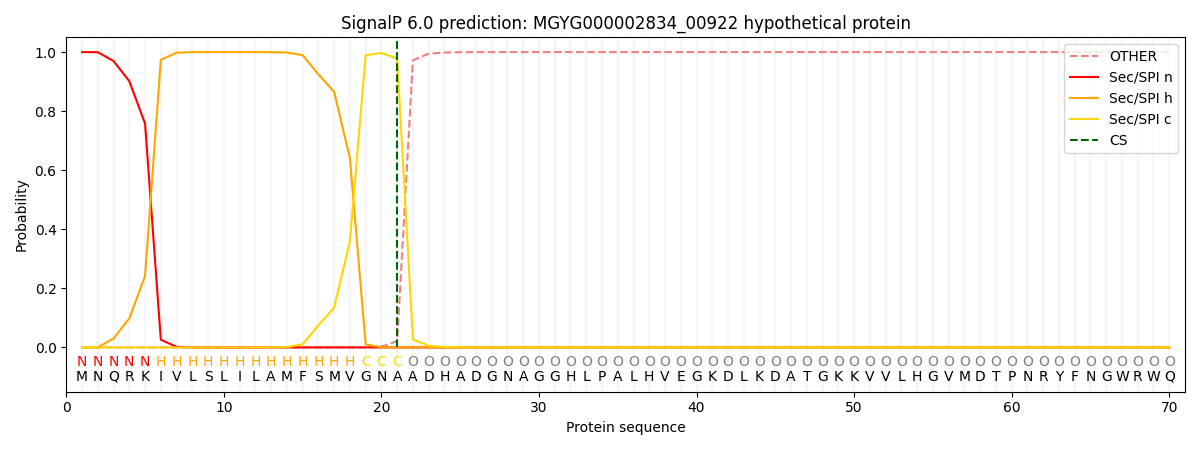
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000289
|
0.998854
|
0.000207
|
0.000227
|
0.000212
|
0.000185
|
There is no transmembrane helices in MGYG000002834_00922.

