You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002762_01544
You are here: Home > Sequence: MGYG000002762_01544
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900554845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900554845 | |||||||||||
| CAZyme ID | MGYG000002762_01544 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10065; End: 12767 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 408 | 552 | 2.9e-19 | 0.7180851063829787 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 9.06e-16 | 697 | 840 | 4 | 150 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam14200 | RicinB_lectin_2 | 2.32e-12 | 444 | 533 | 1 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.91e-10 | 403 | 491 | 7 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 7.05e-10 | 499 | 553 | 3 | 61 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 5.03e-06 | 462 | 546 | 1 | 84 | Ricin-type beta-trefoil lectin domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ60208.1 | 2.03e-188 | 37 | 888 | 49 | 891 |
| QUT88791.1 | 6.27e-187 | 37 | 850 | 49 | 851 |
| ASB47762.1 | 1.12e-185 | 17 | 844 | 26 | 838 |
| AGB29926.1 | 6.52e-159 | 5 | 873 | 6 | 881 |
| QIM09902.1 | 4.56e-155 | 53 | 883 | 28 | 864 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
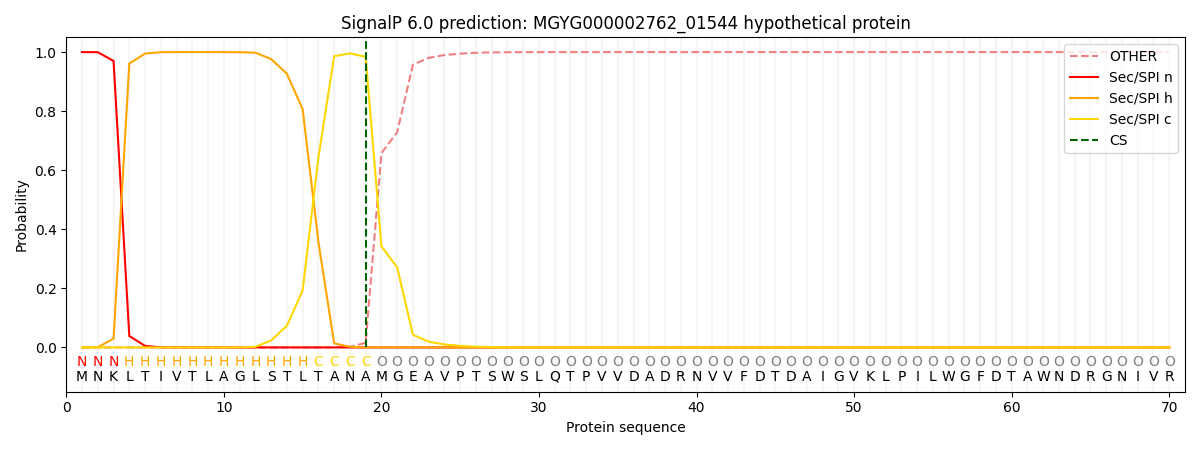
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000327 | 0.998837 | 0.000212 | 0.000225 | 0.000202 | 0.000174 |
