You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002738_00173
You are here: Home > Sequence: MGYG000002738_00173
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
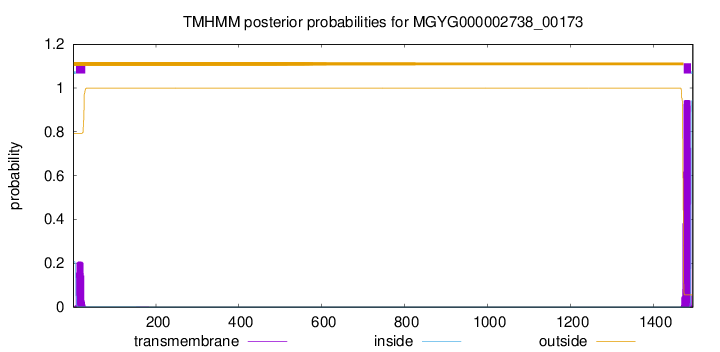
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia; | |||||||||||
| CAZyme ID | MGYG000002738_00173 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2809; End: 7299 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 658 | 1222 | 1.4e-156 | 0.9959266802443992 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 4.32e-10 | 844 | 1001 | 9 | 133 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 3.81e-05 | 794 | 979 | 4 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam18998 | Flg_new_2 | 4.58e-05 | 592 | 651 | 11 | 71 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
| pfam07523 | Big_3 | 3.40e-04 | 1248 | 1315 | 2 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| cd05865 | IgI_1_NCAM-1 | 5.79e-04 | 1354 | 1440 | 3 | 97 | First immunoglobulin (Ig)-like domain of neural cell adhesion molecule (NCAM-1); member of the I-set of Ig superfamily (IgSF) domains. The members here are composed of the first immunoglobulin (Ig)-like domain of neural cell adhesion molecule (NCAM-1). NCAM-1 plays important roles in the development and regeneration of the central nervous system, in synaptogenesis and neural migration. NCAM mediates cell-cell and cell-substratum recognition and adhesion via homophilic (NCAM-NCAM), and heterophilic (NCAM-nonNCAM), interactions. NCAM is expressed as three major isoforms having different intracellular extensions. The extracellular portion of NCAM has five N-terminal Ig-like domains and two fibronectin type III domains. The double zipper adhesion complex model for NCAM homophilic binding involves the Ig1, Ig2, and Ig3 domains. By this model, Ig1 and Ig2 mediate dimerization of NCAM molecules situated on the same cell surface (cis interactions), and Ig3 domains mediate interactions between NCAM molecules expressed on the surface of opposing cells (trans interactions), through binding to the Ig1 and Ig2 domains. The adhesive ability of NCAM is modulated by the addition of polysialic acid chains to the fifth Ig-like domain. This group belongs to the I-set of IgSF domains, having A-B-E-D strands in one beta-sheet and A'-G-F-C-C' in the other. Like the V-set Ig domains, members of the I-set have a discontinuous A strand, but lack a C" strand. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIY03209.1 | 2.08e-284 | 120 | 1229 | 29 | 1169 |
| SDS97815.1 | 5.99e-276 | 123 | 1228 | 51 | 1190 |
| QLL50162.1 | 1.74e-205 | 309 | 1224 | 47 | 979 |
| QLL52630.1 | 1.74e-205 | 309 | 1224 | 47 | 979 |
| ALA27928.1 | 1.74e-205 | 309 | 1224 | 47 | 979 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 7.09e-62 | 657 | 1163 | 7 | 566 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 1.98e-54 | 658 | 1153 | 17 | 571 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 2.53e-50 | 658 | 1222 | 13 | 608 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 3.76e-42 | 658 | 1005 | 244 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 8.97e-42 | 658 | 1005 | 244 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
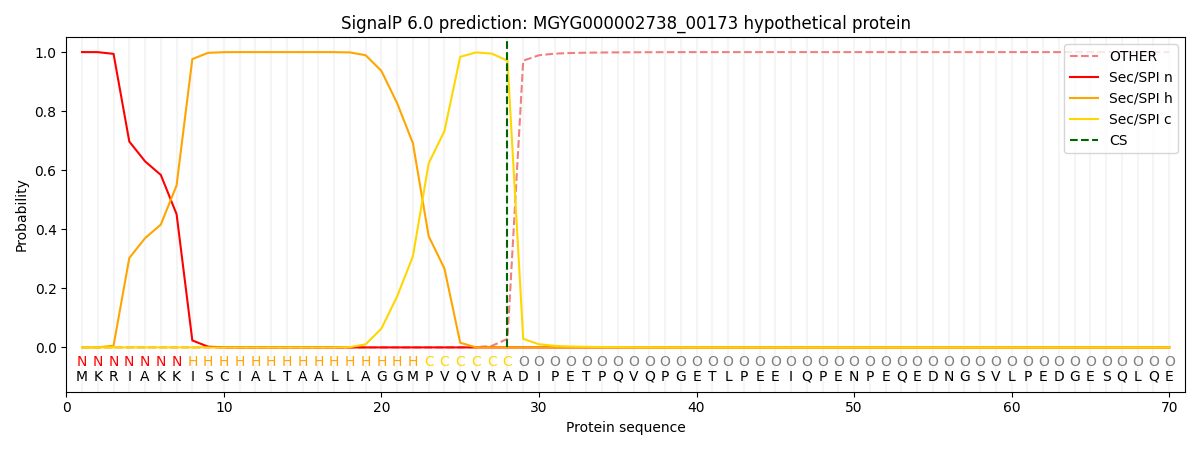
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000245 | 0.999079 | 0.000200 | 0.000169 | 0.000157 | 0.000132 |