You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002707_01054
You are here: Home > Sequence: MGYG000002707_01054
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
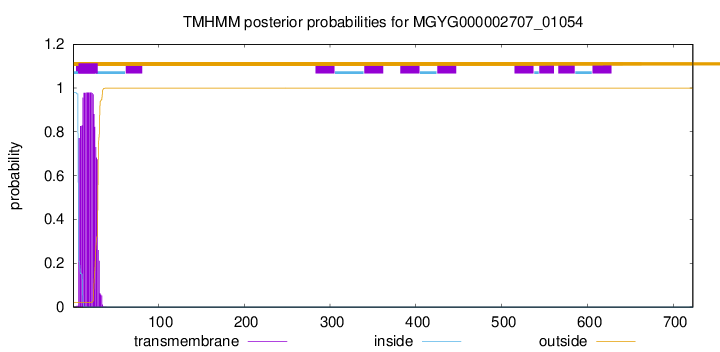
TMHMM annotations
Basic Information help
| Species | Flavonifractor sp900549795 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Flavonifractor; Flavonifractor sp900549795 | |||||||||||
| CAZyme ID | MGYG000002707_01054 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 46267; End: 48438 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 236 | 513 | 8.6e-70 | 0.9923954372623575 |
| CBM32 | 594 | 714 | 1.4e-18 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.51e-21 | 255 | 512 | 22 | 268 | Cellulase (glycosyl hydrolase family 5). |
| pfam00754 | F5_F8_type_C | 1.74e-15 | 593 | 714 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| COG2730 | BglC | 8.50e-15 | 229 | 457 | 42 | 248 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG3934 | COG3934 | 8.07e-05 | 267 | 416 | 37 | 179 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| cd00057 | FA58C | 0.003 | 609 | 697 | 29 | 126 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ99063.1 | 8.20e-133 | 31 | 721 | 36 | 691 |
| SDU77934.1 | 5.02e-110 | 54 | 714 | 321 | 945 |
| SDS04671.1 | 3.40e-102 | 55 | 576 | 52 | 566 |
| ABX04818.1 | 3.04e-93 | 55 | 586 | 54 | 556 |
| QYF74471.1 | 5.82e-92 | 114 | 716 | 360 | 941 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EQP_A | 2.79e-12 | 228 | 448 | 44 | 251 | Exo-b-(1,3)-glucanaseFrom Candida Albicans [Candida albicans] |
| 3O6A_A | 5.07e-12 | 228 | 477 | 49 | 288 | F144Y/F258YDouble Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A [Candida albicans] |
| 4M80_A | 6.74e-12 | 228 | 469 | 49 | 293 | Thestructure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution [Candida albicans SC5314],4M81_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution [Candida albicans SC5314],4M82_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with p-nitrophenyl-gentiobioside (product) at 1.6A resolution [Candida albicans SC5314] |
| 2PC8_A | 6.78e-12 | 228 | 479 | 50 | 297 | ChainA, Hypothetical protein XOG1 [Candida albicans] |
| 1CZ1_A | 8.73e-12 | 228 | 448 | 44 | 251 | Exo-b-(1,3)-glucanaseFrom Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 2.84e-22 | 229 | 516 | 43 | 341 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| Q0C8Z0 | 9.21e-18 | 241 | 532 | 84 | 370 | Probable glucan endo-1,6-beta-glucosidase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=exgB PE=3 SV=1 |
| B8NBJ4 | 3.65e-15 | 237 | 486 | 78 | 325 | Probable glucan endo-1,6-beta-glucosidase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgB PE=3 SV=1 |
| Q2TZQ9 | 4.07e-15 | 237 | 486 | 78 | 325 | Probable glucan endo-1,6-beta-glucosidase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=exgB PE=3 SV=1 |
| B0XRX9 | 2.13e-14 | 237 | 520 | 77 | 356 | Probable glucan endo-1,6-beta-glucosidase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
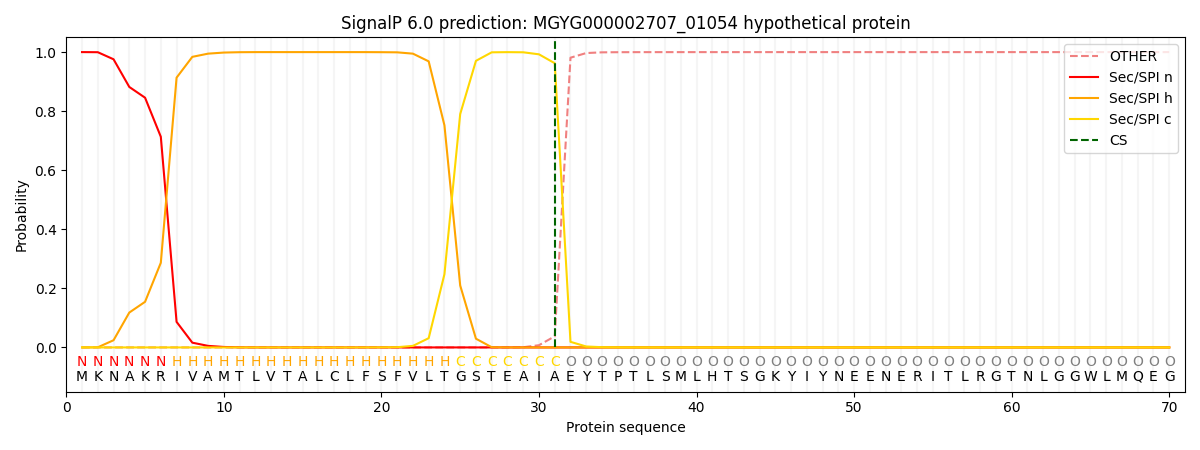
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000232 | 0.999033 | 0.000185 | 0.000197 | 0.000167 | 0.000148 |