You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002581_00783
You are here: Home > Sequence: MGYG000002581_00783
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-269 sp001916065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; CAG-269; CAG-269 sp001916065 | |||||||||||
| CAZyme ID | MGYG000002581_00783 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37898; End: 40723 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 41 | 202 | 2.5e-26 | 0.8031914893617021 |
| CBM13 | 196 | 289 | 2.4e-21 | 0.4787234042553192 |
| CBM13 | 353 | 444 | 7.3e-17 | 0.46808510638297873 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14200 | RicinB_lectin_2 | 3.59e-27 | 234 | 322 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 7.23e-27 | 393 | 479 | 3 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 3.46e-26 | 78 | 166 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| COG4193 | LytD | 1.59e-25 | 568 | 780 | 45 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam14200 | RicinB_lectin_2 | 7.39e-21 | 195 | 275 | 10 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM02197.1 | 2.98e-44 | 493 | 894 | 148 | 561 |
| ACR76042.1 | 4.98e-42 | 491 | 787 | 229 | 533 |
| CBK91761.1 | 8.84e-42 | 491 | 787 | 229 | 533 |
| CBK92679.1 | 1.57e-41 | 491 | 787 | 229 | 533 |
| QUI95360.1 | 5.23e-40 | 508 | 783 | 53 | 342 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6U0O_B | 2.82e-18 | 568 | 773 | 73 | 269 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 6FXP_A | 5.96e-18 | 568 | 773 | 43 | 239 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 1.28e-11 | 571 | 781 | 37 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 3.98e-09 | 638 | 775 | 96 | 226 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 2.38e-08 | 638 | 775 | 96 | 226 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6GAG0 | 5.42e-10 | 571 | 781 | 1043 | 1250 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q8NX96 | 5.43e-10 | 571 | 781 | 1049 | 1256 | Bifunctional autolysin OS=Staphylococcus aureus (strain MW2) OX=196620 GN=atl PE=3 SV=1 |
| Q931U5 | 7.11e-10 | 571 | 781 | 1041 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q99V41 | 7.11e-10 | 571 | 781 | 1041 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| A7X0T9 | 7.13e-10 | 571 | 781 | 1048 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu3 / ATCC 700698) OX=418127 GN=atl PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
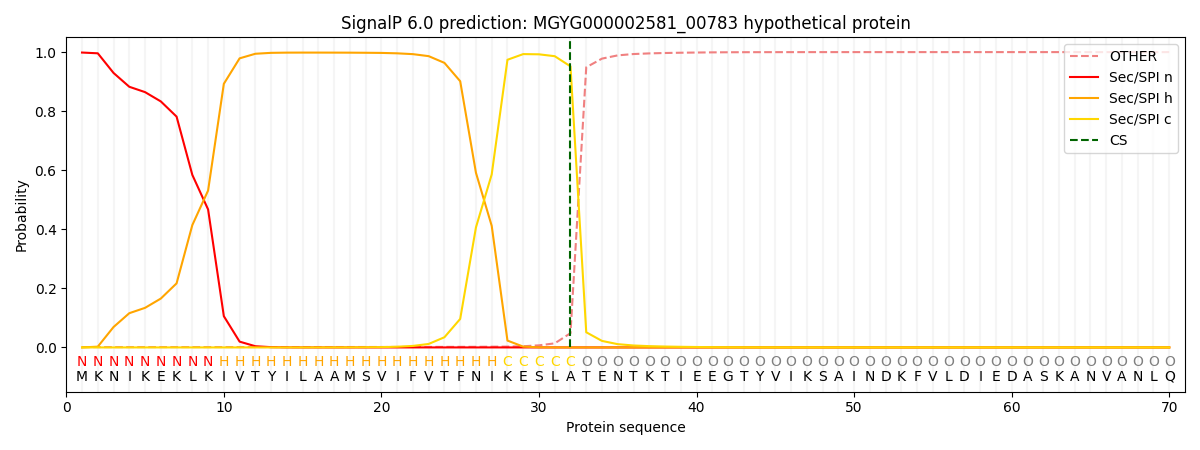
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002295 | 0.996500 | 0.000597 | 0.000204 | 0.000179 | 0.000184 |
