You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002473_03227
You are here: Home > Sequence: MGYG000002473_03227
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis vadensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis vadensis | |||||||||||
| CAZyme ID | MGYG000002473_03227 | |||||||||||
| CAZy Family | GH167 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 480; End: 3737 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH167 | 379 | 845 | 9.5e-44 | 0.6317991631799164 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd03143 | A4_beta-galactosidase_middle_domain | 5.48e-09 | 664 | 825 | 2 | 147 | A4 beta-galactosidase middle domain: a type 1 glutamine amidotransferase (GATase1)-like domain. A4 beta-galactosidase middle domain: a type 1 glutamine amidotransferase (GATase1)-like domain. This group includes proteins similar to beta-galactosidase from Thermus thermophilus. Beta-Galactosidase hydrolyzes the beta-1,4-D-galactosidic linkage of lactose, as well as those of related chromogens, o-nitrophenyl-beta-D-galactopyranoside (ONP-Gal) and 5-bromo-4-chloro-3-indolyl-beta-D-galactoside (X-gal). This A4 beta-galactosidase middle domain lacks the catalytic triad of typical GATase1 domains. The reactive Cys residue found in the sharp turn between a beta strand and an alpha helix termed the nucleophile elbow in typical GATase1 domains is not conserved in this group. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46758.1 | 0.0 | 16 | 1067 | 15 | 1061 |
| AVM45830.1 | 5.83e-33 | 193 | 1062 | 38 | 922 |
| AVM45809.1 | 7.02e-23 | 959 | 1069 | 1005 | 1118 |
| AVM46610.1 | 4.16e-21 | 960 | 1069 | 987 | 1099 |
| AVM46955.1 | 7.19e-21 | 970 | 1069 | 1011 | 1112 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PTM_A | 9.57e-13 | 406 | 795 | 316 | 687 | Crystalstructure of apo exo-carrageenase GH42 from Bacteroides ovatus [Bacteroides ovatus CL02T12C04] |
| 4OIF_A | 3.64e-10 | 403 | 762 | 169 | 509 | 3Dstructure of Gan42B, a GH42 beta-galactosidase from G. [Geobacillus stearothermophilus],4OIF_B 3D structure of Gan42B, a GH42 beta-galactosidase from G. [Geobacillus stearothermophilus],4OIF_C 3D structure of Gan42B, a GH42 beta-galactosidase from G. [Geobacillus stearothermophilus] |
| 4OJY_A | 3.64e-10 | 403 | 762 | 170 | 510 | 3Dstructure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus [Geobacillus stearothermophilus],4OJY_B 3D structure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus [Geobacillus stearothermophilus],4OJY_C 3D structure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus [Geobacillus stearothermophilus] |
| 5DFA_A | 1.09e-09 | 403 | 762 | 169 | 509 | 3Dstructure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus [Geobacillus stearothermophilus],5DFA_B 3D structure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus [Geobacillus stearothermophilus],5DFA_C 3D structure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q65CX4 | 9.66e-11 | 406 | 811 | 158 | 551 | Beta-galactosidase GalA OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=lacA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
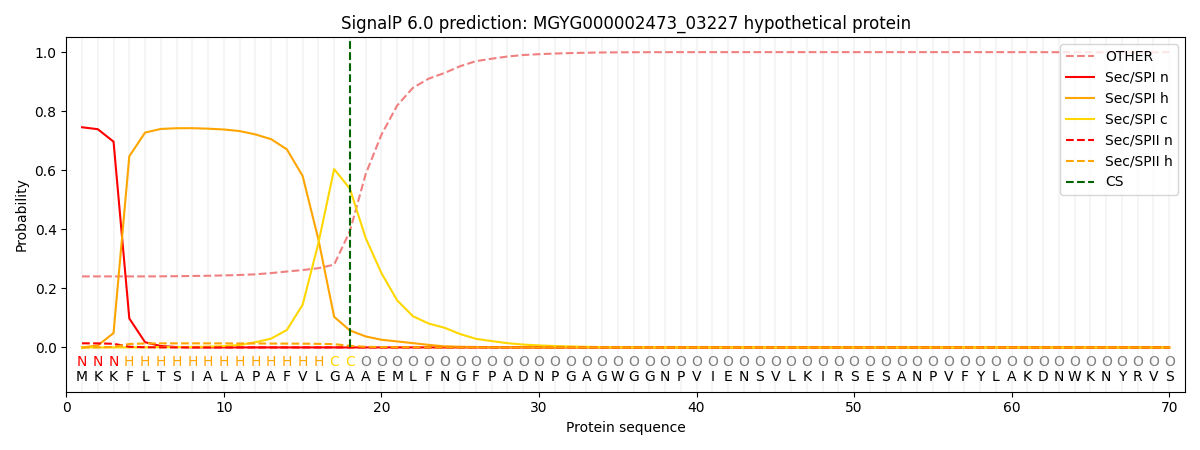
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.249696 | 0.734344 | 0.015032 | 0.000311 | 0.000273 | 0.000317 |
