You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002459_01455
You are here: Home > Sequence: MGYG000002459_01455
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bifidobacterium animalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium animalis | |||||||||||
| CAZyme ID | MGYG000002459_01455 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Mannan endo-1,4-beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1703417; End: 1704553 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 44 | 290 | 1.4e-93 | 0.9802371541501976 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.02e-34 | 43 | 286 | 3 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 3.62e-06 | 70 | 299 | 82 | 343 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam02013 | CBM_10 | 2.87e-05 | 351 | 375 | 8 | 34 | Cellulose or protein binding domain. This domain is found in two distinct sets of proteins with different functions. Those found in aerobic bacteria bind cellulose (or other carbohydrates); but in anaerobic fungi they are protein binding domains, referred to as dockerin domains or docking domains. They are believed to be responsible for the assembly of a multiprotein cellulase/hemicellulase complex, similar to the cellulosome found in certain anaerobic bacteria. |
| smart01064 | CBM_10 | 2.05e-04 | 350 | 375 | 7 | 27 | Cellulose or protein binding domain. This domain is found in two distinct sets of proteins with different functions. Those found in aerobic bacteria bind cellulose (or other carbohydrates); but in anaerobic fungi they are protein binding domains, referred to as dockerin domains or docking domains. They are believed to be responsible for the assembly of a multiprotein cellulase/hemicellulase complex, similar to the cellulosome found in certain anaerobic bacteria. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIA33432.1 | 4.64e-277 | 1 | 378 | 1 | 378 |
| AJD89158.1 | 4.64e-277 | 1 | 378 | 1 | 378 |
| QRI03701.1 | 4.64e-277 | 1 | 378 | 1 | 378 |
| AYD77908.1 | 4.64e-277 | 1 | 378 | 1 | 378 |
| AXM92966.1 | 4.64e-277 | 1 | 378 | 1 | 378 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1BQC_A | 1.05e-105 | 34 | 330 | 4 | 302 | Beta-MannanaseFrom Thermomonospora Fusca [Thermobifida fusca],2MAN_A Mannotriose Complex Of Thermomonospora Fusca Beta-Mannanase [Thermobifida fusca],3MAN_A Mannohexaose Complex Of Thermomonospora Fusca Beta-mannanase [Thermobifida fusca] |
| 4FK9_A | 3.08e-95 | 34 | 332 | 24 | 321 | HighResolution Structure of the Catalytic Domain of Mannanase SActE_2347 from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 3WSU_A | 1.36e-90 | 29 | 318 | 34 | 323 | Crystalstructure of beta-mannanase from Streptomyces thermolilacinus [Streptomyces thermolilacinus],3WSU_B Crystal structure of beta-mannanase from Streptomyces thermolilacinus [Streptomyces thermolilacinus] |
| 4Y7E_A | 1.09e-89 | 29 | 318 | 34 | 323 | Crystalstructure of beta-mannanase from Streptomyces thermolilacinus with mannohexaose [Streptomyces thermolilacinus],4Y7E_B Crystal structure of beta-mannanase from Streptomyces thermolilacinus with mannohexaose [Streptomyces thermolilacinus] |
| 1WKY_A | 4.40e-76 | 30 | 333 | 6 | 308 | Crystalstructure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module [Bacillus sp. JAMB-602] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P51529 | 6.95e-126 | 11 | 378 | 17 | 377 | Mannan endo-1,4-beta-mannosidase OS=Streptomyces lividans OX=1916 GN=manA PE=1 SV=2 |
| B3PF24 | 5.65e-102 | 26 | 377 | 41 | 389 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=man5A PE=1 SV=1 |
| P22533 | 2.88e-84 | 52 | 320 | 55 | 325 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
| G1K3N4 | 8.53e-67 | 35 | 319 | 4 | 287 | Mannan endo-1,4-beta-mannosidase OS=Salipaludibacillus agaradhaerens OX=76935 PE=1 SV=1 |
| P54937 | 7.82e-13 | 67 | 281 | 86 | 342 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
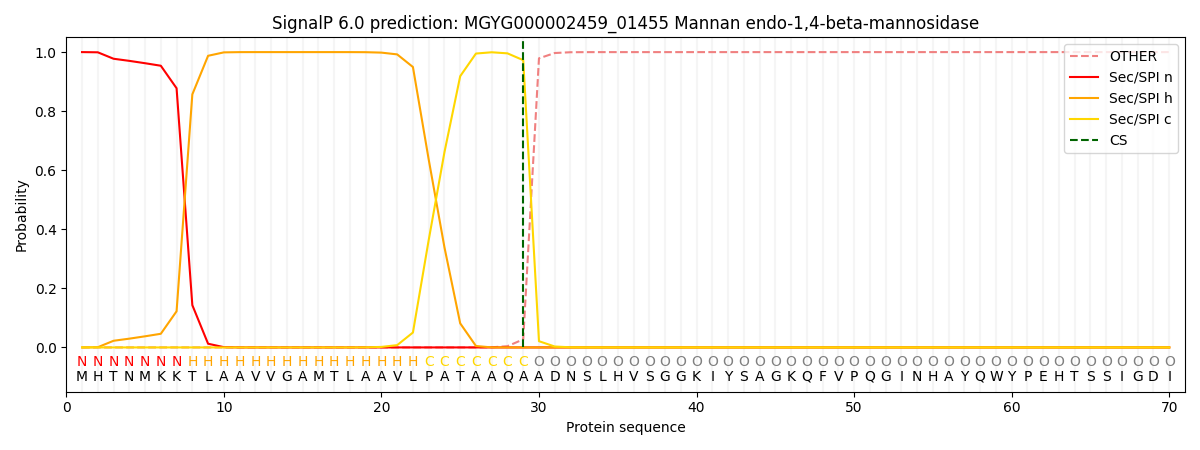
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.998997 | 0.000186 | 0.000208 | 0.000188 | 0.000155 |
