You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002455_00685
You are here: Home > Sequence: MGYG000002455_00685
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
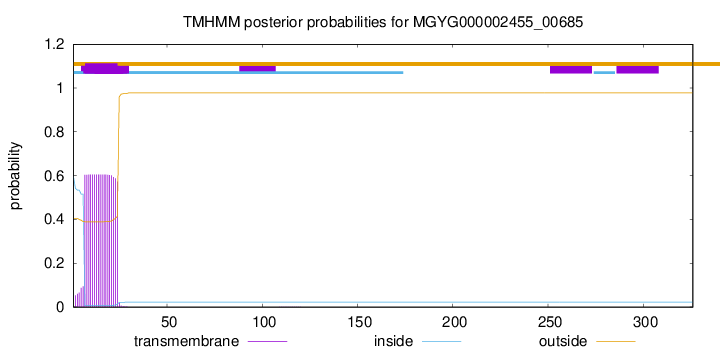
TMHMM annotations
Basic Information help
| Species | Bacteroides cellulosilyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides cellulosilyticus | |||||||||||
| CAZyme ID | MGYG000002455_00685 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 398422; End: 399402 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 50 | 283 | 7.9e-94 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.33e-55 | 47 | 291 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 6.20e-06 | 112 | 288 | 118 | 360 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd06329 | PBP1_SBP-like | 9.43e-05 | 72 | 195 | 201 | 314 | periplasmic substrate-binding domain of active transport proteins (substrate binding proteins or SBPs). Periplasmic substrate-binding domain of active transport proteins found in bacteria and Archaea. Members of this group are initial receptors in the process of active transport across cellular membrane, but their substrate specificities are not known in detail. However, they closely resemble the group of AmiC and active transport systems for short-chain amides and urea (FmdDEF), and thus are likely to exhibit a ligand-binding mode similar to that of the amide sensor protein AmiC from Pseudomonas aeruginosa. Moreover, this binding domain has high sequence identity to the family of hydrophobic amino acid transporters (HAAT), and thus it may also be involved in transport of amino acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT89667.1 | 3.11e-251 | 1 | 326 | 1 | 326 |
| ALJ59275.1 | 6.48e-244 | 1 | 326 | 1 | 326 |
| QUU00598.1 | 2.35e-202 | 2 | 325 | 3 | 323 |
| QUT65069.1 | 7.52e-202 | 2 | 323 | 3 | 321 |
| SIP56247.1 | 7.52e-202 | 2 | 323 | 3 | 321 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 1.98e-79 | 38 | 325 | 12 | 302 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZW_A | 1.22e-77 | 38 | 323 | 11 | 302 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 1.01e-76 | 38 | 323 | 11 | 303 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 5IHS_A | 2.44e-74 | 12 | 326 | 9 | 323 | Structureof CHU_2103 from Cytophaga hutchinsonii [Cytophaga hutchinsonii ATCC 33406] |
| 3PZT_A | 2.70e-74 | 38 | 323 | 36 | 324 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10475 | 4.69e-74 | 6 | 323 | 6 | 329 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 1.31e-73 | 6 | 323 | 6 | 329 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P06565 | 7.76e-73 | 12 | 325 | 13 | 328 | Endoglucanase B OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celB PE=3 SV=1 |
| O85465 | 2.40e-72 | 12 | 325 | 13 | 328 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
| P23549 | 1.12e-71 | 6 | 323 | 6 | 329 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
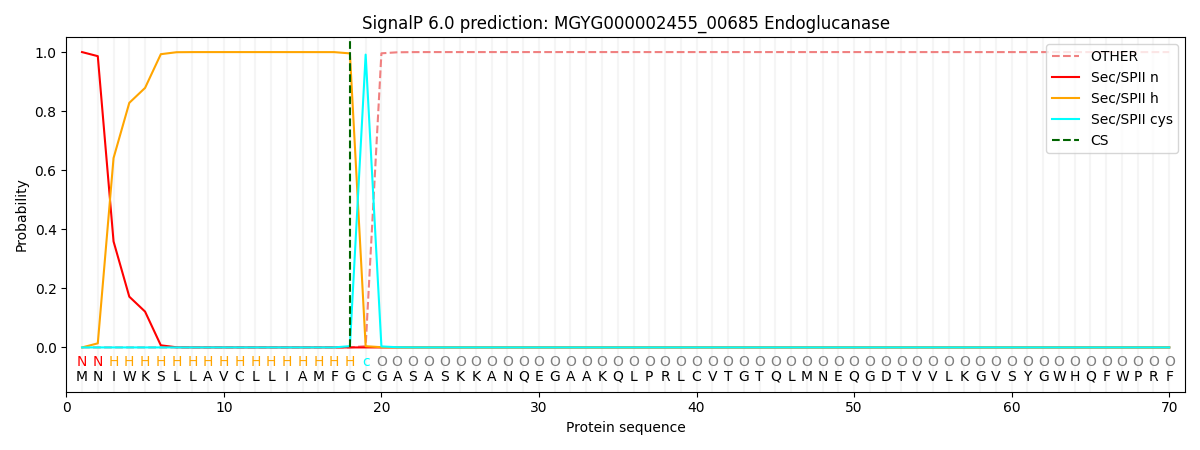
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000069 | 0.000000 | 0.000000 | 0.000000 |