You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002403_00465
You are here: Home > Sequence: MGYG000002403_00465
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
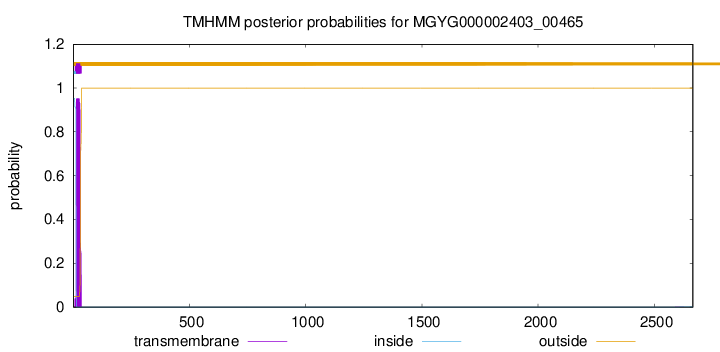
TMHMM annotations
Basic Information help
| Species | Robinsoniella peoriensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella peoriensis | |||||||||||
| CAZyme ID | MGYG000002403_00465 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20585; End: 28585 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 388 | 536 | 2.9e-29 | 0.9850746268656716 |
| GH98 | 1206 | 1504 | 1.6e-16 | 0.9113149847094801 |
| CBM32 | 1958 | 2089 | 2.1e-16 | 0.9516129032258065 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 5.45e-26 | 385 | 537 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08307 | Glyco_hydro_98C | 4.77e-23 | 1521 | 1775 | 1 | 269 | Glycosyl hydrolase family 98 C-terminal domain. This putative domain is found at the C-terminus of glycosyl hydrolase family 98 proteins. This domain is not expected to form part of the catalytic activity. |
| smart00776 | NPCBM | 5.66e-22 | 384 | 536 | 2 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| COG5492 | YjdB | 9.29e-19 | 2144 | 2356 | 144 | 329 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 9.91e-19 | 2312 | 2505 | 127 | 318 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQR05375.1 | 2.70e-165 | 715 | 1201 | 22 | 503 |
| ANU41753.1 | 2.70e-165 | 715 | 1201 | 22 | 503 |
| QIA30400.1 | 3.69e-165 | 715 | 1201 | 22 | 503 |
| QIG40118.1 | 4.52e-92 | 1183 | 1773 | 20 | 566 |
| CBK83841.1 | 8.35e-46 | 2423 | 2666 | 1535 | 1782 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6N1B_A | 2.73e-170 | 719 | 1201 | 27 | 497 | Crystalstructure of an N-acetylgalactosamine deacetylase from F. plautii in complex with blood group B trisaccharide [Flavonifractor plautii] |
| 6N1A_A | 6.24e-169 | 719 | 1201 | 27 | 497 | Crystalstructure of an N-acetylgalactosamine deacetylase from F. plautii [Flavonifractor plautii] |
| 2WMH_A | 6.11e-43 | 1204 | 1773 | 26 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 in complex with the H- disaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
| 2WMF_A | 2.67e-42 | 1204 | 1773 | 26 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. [Streptococcus pneumoniae TIGR4] |
| 2WMG_A | 3.58e-42 | 1204 | 1773 | 26 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in complex with the LewisY pentasaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR4 | 5.41e-166 | 715 | 1201 | 22 | 503 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| Q0TR53 | 2.41e-17 | 1832 | 2116 | 527 | 782 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 7.12e-17 | 1832 | 2091 | 527 | 765 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| P33747 | 6.09e-14 | 2195 | 2352 | 37 | 191 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| P85991 | 2.13e-13 | 2277 | 2445 | 84 | 257 | Ig-like virion protein OS=Serratia phage KSP90 OX=552528 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
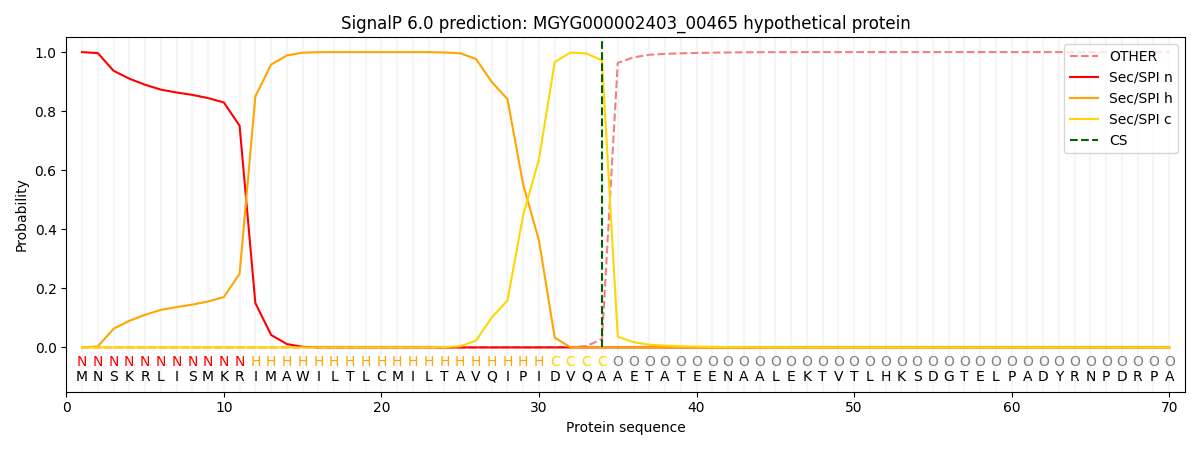
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000256 | 0.999064 | 0.000198 | 0.000160 | 0.000152 | 0.000141 |