You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002371_00516
You are here: Home > Sequence: MGYG000002371_00516
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Tidjanibacter inops_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Tidjanibacter; Tidjanibacter inops_A | |||||||||||
| CAZyme ID | MGYG000002371_00516 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5630; End: 7648 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 17 | 552 | 7.6e-82 | 0.586436170212766 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.12e-36 | 13 | 398 | 9 | 403 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.33e-33 | 14 | 475 | 10 | 496 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 8.63e-25 | 59 | 428 | 111 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02836 | Glyco_hydro_2_C | 2.70e-13 | 282 | 465 | 8 | 193 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 2.02e-09 | 13 | 128 | 13 | 135 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG53562.1 | 1.33e-201 | 1 | 643 | 15 | 657 |
| QGA24357.1 | 3.26e-201 | 1 | 643 | 7 | 651 |
| CDN31448.1 | 1.17e-169 | 2 | 645 | 8 | 657 |
| QUT71232.1 | 6.41e-169 | 2 | 645 | 16 | 665 |
| QQA06349.1 | 9.06e-169 | 1 | 648 | 15 | 668 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CMG_A | 2.99e-161 | 32 | 645 | 24 | 639 | Crystalstructure of putative beta-galactosidase from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 5Z1A_A | 4.47e-161 | 32 | 645 | 43 | 658 | Thecrystal structure of Bacteroides fragilis beta-glucuronidase in complex with uronic isofagomine [Bacteroides fragilis NCTC 9343] |
| 6MVG_A | 6.15e-109 | 12 | 624 | 23 | 624 | Crystalstructure of FMN-binding beta-glucuronidase from Ruminococcus gnavus [[Ruminococcus] gnavus],6MVG_B Crystal structure of FMN-binding beta-glucuronidase from Ruminococcus gnavus [[Ruminococcus] gnavus],6MVG_C Crystal structure of FMN-binding beta-glucuronidase from Ruminococcus gnavus [[Ruminococcus] gnavus] |
| 6MVH_A | 2.27e-97 | 36 | 661 | 54 | 659 | Crystalstructure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis],6MVH_B Crystal structure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis],6MVH_C Crystal structure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis],6MVH_D Crystal structure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis] |
| 7KGZ_A | 2.10e-90 | 14 | 660 | 2 | 623 | ChainA, Beta-glucuronidase [Roseburia hominis],7KGZ_B Chain B, Beta-glucuronidase [Roseburia hominis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 2.51e-48 | 36 | 667 | 82 | 721 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| P77989 | 1.01e-34 | 13 | 449 | 2 | 431 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| A7LXS9 | 5.01e-28 | 2 | 398 | 30 | 434 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| T2KM09 | 7.71e-27 | 36 | 620 | 78 | 665 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| T2KN75 | 3.00e-26 | 14 | 640 | 26 | 664 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
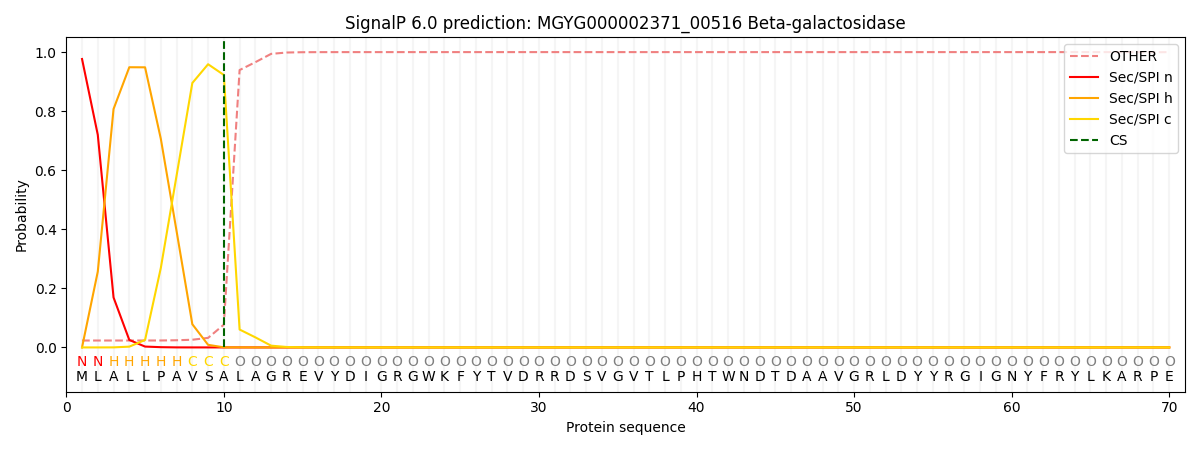
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.025522 | 0.973314 | 0.000279 | 0.000340 | 0.000275 | 0.000256 |
