You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002370_03145
You are here: Home > Sequence: MGYG000002370_03145
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridioides difficile_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Clostridioides; Clostridioides difficile_A | |||||||||||
| CAZyme ID | MGYG000002370_03145 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43899; End: 45485 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033435 | S-layer_Clost | 3.57e-31 | 21 | 277 | 376 | 693 | S-layer protein SlpA. In Clostridiodes difficile, the S-layer protein precursor, SlpA, is one member of a large paralogous family of protein that share several cell wall-binding repeats. SlpA is cleaved into a larger and smaller protein. The S-layer protein itself is important to adhesion, and portions of it are highly variable, and then N-terminal and C-terminal are well-conserved. |
| cd05379 | CAP_bacterial | 9.71e-27 | 397 | 525 | 1 | 119 | Bacterial CAP (cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins) domain proteins. Little is known about bacterial and archaeal members of the CAP (cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins) domain family. The wider family of CAP domain containing proteins includes plant pathogenesis-related protein 1 (PR-1), cysteine-rich secretory proteins (CRISPs), and allergen 5 from vespid venom, among others. Studies of eukaryotic proteins show that CAP domains have several functions, including the binding of cholesterol, lipids and heparan sulfate. This group includes Borrelia burgdorferi outer surface protein BB0689, which does not bind to cholesterol, lipids, or heparan sulfate, and whose function is unknown. |
| pfam04122 | CW_binding_2 | 5.49e-19 | 33 | 114 | 2 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| pfam04122 | CW_binding_2 | 1.47e-14 | 128 | 210 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| pfam04122 | CW_binding_2 | 7.79e-14 | 219 | 289 | 1 | 73 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQY46700.1 | 2.14e-105 | 1 | 310 | 1 | 308 |
| AVD38579.1 | 2.14e-105 | 1 | 310 | 1 | 308 |
| AVD42108.1 | 2.14e-105 | 1 | 310 | 1 | 308 |
| QQY76565.1 | 2.14e-105 | 1 | 310 | 1 | 308 |
| AFV69567.1 | 2.14e-105 | 1 | 310 | 1 | 308 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J6Q_A | 9.63e-36 | 30 | 297 | 286 | 586 | ChainA, Cell wall binding protein cwp8 [Clostridioides difficile 630] |
| 5J72_A | 1.11e-34 | 30 | 310 | 132 | 441 | ChainA, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630],5J72_B Chain B, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630] |
| 7ACZ_B | 1.28e-30 | 25 | 282 | 35 | 354 | ChainB, SLPH (HMW SLP) [Clostridioides difficile R20291],7ACZ_D Chain D, SLPH (HMW SLP) [Clostridioides difficile R20291] |
| 7ACX_B | 1.10e-29 | 25 | 277 | 35 | 338 | ChainB, S-layer protein [Clostridioides difficile],7ACX_D Chain D, S-layer protein [Clostridioides difficile] |
| 7ACY_B | 3.83e-29 | 25 | 277 | 35 | 338 | ChainB, S-layer protein [Clostridioides difficile 630],7ACY_D Chain D, S-layer protein [Clostridioides difficile 630],7QGQ_B Chain B, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_D Chain D, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_J Chain J, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_K Chain K, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_L Chain L, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_M Chain M, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_N Chain N, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_T Chain T, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_U Chain U, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_V Chain V, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_W Chain W, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_X Chain X, Precursor of the S-layer proteins [Clostridioides difficile 630] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02114 | 4.23e-24 | 28 | 273 | 26 | 279 | N-acetylmuramoyl-L-alanine amidase LytC OS=Bacillus subtilis (strain 168) OX=224308 GN=lytC PE=1 SV=1 |
| Q02113 | 4.70e-13 | 24 | 248 | 154 | 351 | Amidase enhancer OS=Bacillus subtilis (strain 168) OX=224308 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
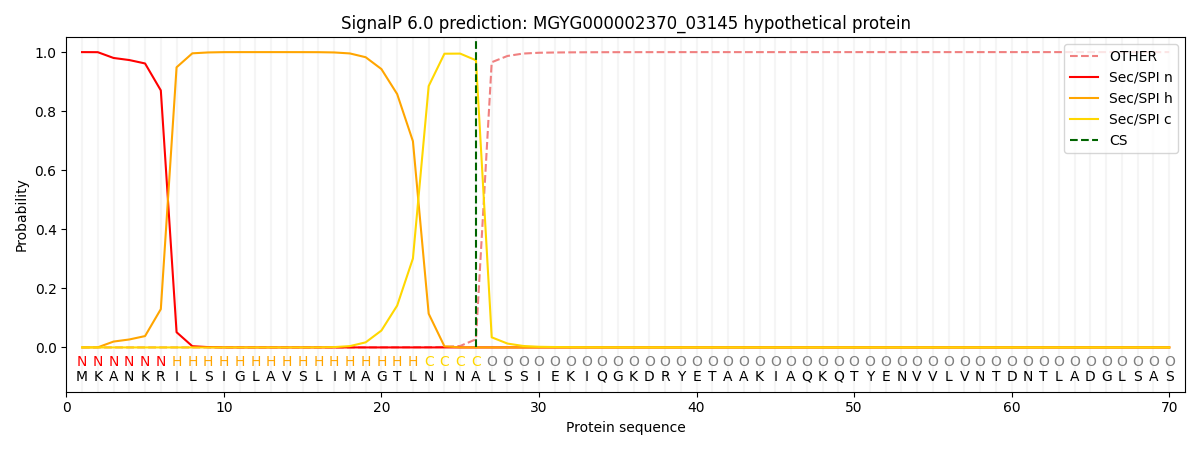
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000300 | 0.998959 | 0.000219 | 0.000174 | 0.000166 | 0.000149 |

