You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002367_01431
You are here: Home > Sequence: MGYG000002367_01431
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pediococcus pentosaceus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Pediococcus; Pediococcus pentosaceus | |||||||||||
| CAZyme ID | MGYG000002367_01431 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 127222; End: 127908 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13925 | RPF | 1.00e-12 | 167 | 227 | 1 | 69 | core lysozyme-like domain of resuscitation-promoting factor proteins. Resuscitation-promoting factor (RPF) proteins, found in various (G+C)-rich Gram-positive bacteria, act to reactivate cultures from stationary phase. This protein shares elements of the structural core of lysozyme and related proteins. Furthermore, it shares a conserved active site glutamate which is required for activity, and has a polysaccharide binding cleft that corresponds to the peptidoglycan binding cleft of lysozyme. Muralytic activity of Rpf in Micrococcus luteus correlates with resuscitation, supporting a mechanism dependent on cleavage of peptidoglycan by RPF. |
| cd00118 | LysM | 8.37e-12 | 36 | 81 | 2 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| pfam01476 | LysM | 5.43e-11 | 37 | 82 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| smart00257 | LysM | 2.08e-10 | 36 | 81 | 1 | 44 | Lysin motif. |
| PRK11198 | PRK11198 | 1.59e-08 | 25 | 81 | 86 | 145 | LysM domain/BON superfamily protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHO67084.1 | 1.62e-115 | 1 | 228 | 1 | 228 |
| QHM67529.1 | 1.62e-115 | 1 | 228 | 1 | 228 |
| QDZ70682.1 | 1.62e-115 | 1 | 228 | 1 | 228 |
| QHM68317.1 | 1.62e-115 | 1 | 228 | 1 | 228 |
| QHM59809.1 | 1.62e-115 | 1 | 228 | 1 | 228 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4A0G5 | 4.59e-26 | 154 | 226 | 168 | 240 | Probable transglycosylase IsaA OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=isaA PE=3 SV=1 |
| Q8CMZ9 | 2.53e-13 | 155 | 227 | 164 | 234 | Probable transglycosylase IsaA OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=isaA PE=3 SV=1 |
| Q5HL49 | 2.53e-13 | 155 | 227 | 164 | 234 | Probable transglycosylase IsaA OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=isaA PE=3 SV=1 |
| Q2YWD9 | 1.16e-11 | 155 | 227 | 162 | 232 | Probable transglycosylase IsaA OS=Staphylococcus aureus (strain bovine RF122 / ET3-1) OX=273036 GN=isaA PE=3 SV=1 |
| Q2FV52 | 4.15e-11 | 155 | 227 | 162 | 232 | Probable transglycosylase IsaA OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=isaA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
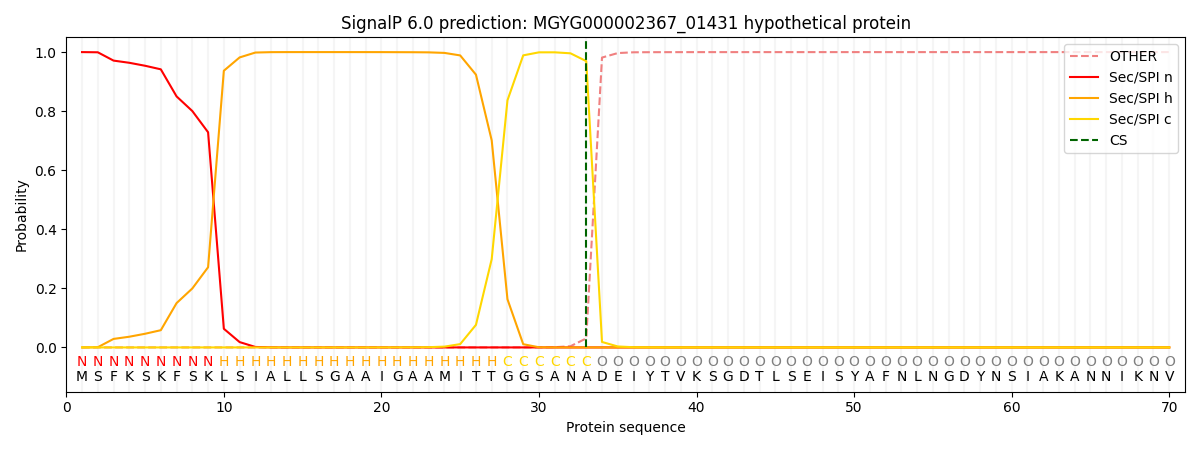
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000297 | 0.999065 | 0.000155 | 0.000182 | 0.000152 | 0.000135 |
