You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002367_01241
You are here: Home > Sequence: MGYG000002367_01241
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pediococcus pentosaceus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Pediococcus; Pediococcus pentosaceus | |||||||||||
| CAZyme ID | MGYG000002367_01241 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 74946; End: 76487 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 5.69e-33 | 411 | 513 | 3 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG0791 | Spr | 2.95e-22 | 411 | 510 | 89 | 195 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| NF033742 | NlpC_p60_RipB | 5.95e-13 | 410 | 498 | 90 | 189 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
| pfam01476 | LysM | 8.30e-13 | 91 | 133 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| NF033741 | NlpC_p60_RipA | 9.48e-12 | 403 | 498 | 334 | 440 | NlpC/P60 family peptidoglycan endopeptidase RipA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDJ24413.1 | 1.35e-167 | 1 | 513 | 1 | 496 |
| ASC08270.1 | 1.55e-166 | 1 | 513 | 1 | 496 |
| QHM65985.1 | 1.55e-166 | 1 | 513 | 1 | 496 |
| QHM67704.1 | 1.55e-166 | 1 | 513 | 1 | 496 |
| ABJ68217.1 | 1.55e-166 | 1 | 513 | 1 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6B8C_A | 3.42e-29 | 411 | 512 | 42 | 142 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 7CFL_A | 8.86e-14 | 397 | 507 | 16 | 129 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 4XCM_A | 1.15e-10 | 410 | 505 | 127 | 220 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
| 2K1G_A | 1.39e-10 | 410 | 490 | 19 | 99 | SolutionNMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 [Escherichia coli K-12] |
| 2HBW_A | 2.35e-07 | 412 | 488 | 115 | 190 | ChainA, NLP/P60 protein [Trichormus variabilis ATCC 29413] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13692 | 1.52e-26 | 411 | 512 | 416 | 516 | Protein P54 OS=Enterococcus faecium OX=1352 PE=3 SV=2 |
| O34669 | 5.35e-15 | 32 | 150 | 23 | 142 | Cell wall-binding protein YocH OS=Bacillus subtilis (strain 168) OX=224308 GN=yocH PE=1 SV=1 |
| P54421 | 9.31e-15 | 32 | 132 | 23 | 129 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| Q49UX4 | 1.23e-13 | 34 | 144 | 26 | 138 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=sle1 PE=3 SV=1 |
| Q5HIL2 | 1.76e-13 | 39 | 148 | 31 | 150 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain COL) OX=93062 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
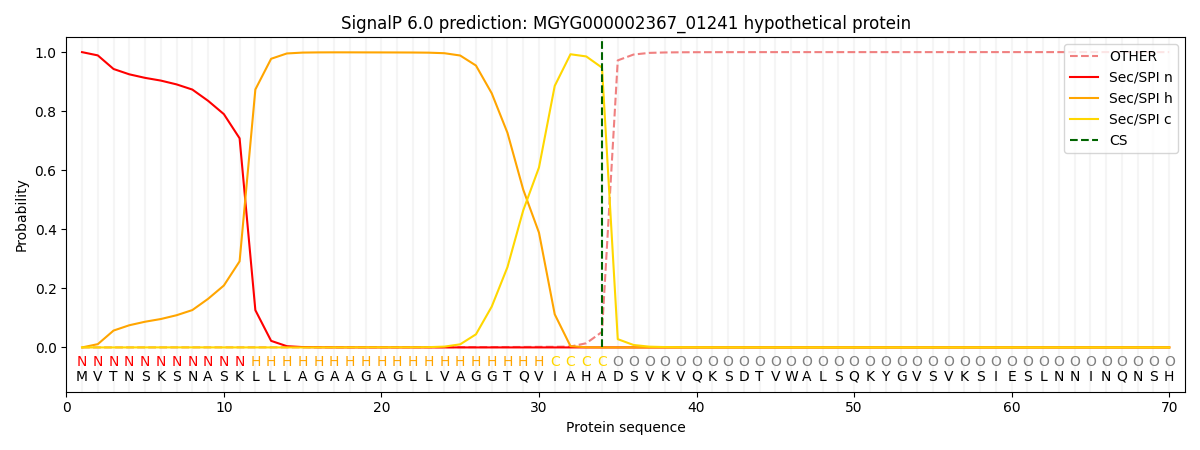
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000480 | 0.998708 | 0.000206 | 0.000246 | 0.000172 | 0.000151 |

