You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002365_01128
You are here: Home > Sequence: MGYG000002365_01128
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
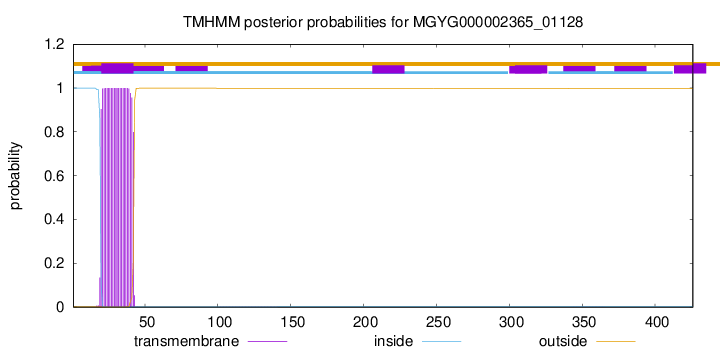
TMHMM annotations
Basic Information help
| Species | Bifidobacterium globosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium globosum | |||||||||||
| CAZyme ID | MGYG000002365_01128 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1253294; End: 1254574 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 158 | 381 | 1.1e-36 | 0.9490740740740741 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 5.71e-49 | 93 | 424 | 1 | 317 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 6.36e-34 | 94 | 418 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 4.04e-17 | 117 | 414 | 18 | 308 | beta-hexosaminidase; Provisional |
| pfam03929 | PepSY_TM | 0.001 | 18 | 161 | 179 | 334 | PepSY-associated TM region. The PepSY_TM family is so named because it is an alignment of up to five transmembranes helices found in bacterial species some of which carry a nested PepSY domain, pfam03413. |
| PRK15098 | PRK15098 | 0.002 | 261 | 424 | 203 | 358 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIZ16452.1 | 7.29e-287 | 1 | 426 | 1 | 426 |
| ATO39883.1 | 1.06e-279 | 1 | 426 | 1 | 426 |
| ASW24369.1 | 4.22e-161 | 180 | 426 | 1 | 247 |
| ATU20885.1 | 1.33e-153 | 83 | 424 | 106 | 447 |
| BAQ31653.1 | 5.92e-147 | 85 | 425 | 76 | 416 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BU9_A | 1.85e-68 | 93 | 419 | 5 | 337 | Crystalstructure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333],5BU9_B Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333] |
| 6K5J_A | 3.72e-27 | 93 | 419 | 11 | 337 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 6GFV_A | 2.37e-20 | 93 | 419 | 16 | 337 | Mtuberculosis LpqI [Mycobacterium tuberculosis H37Rv],6GFV_B M tuberculosis LpqI [Mycobacterium tuberculosis H37Rv] |
| 4GVF_A | 7.22e-19 | 117 | 394 | 18 | 292 | Crystalstructure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to GlcNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVF_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to GlcNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVG_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVG_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVH_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) covalently bound to 5-fluoro-GlcNAc. [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVH_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) covalently bound to 5-fluoro-GlcNAc. [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4HZM_A Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4HZM_B Crystal structure of Salmonella typhimurium family 3 glycoside hydrolase (NagZ) bound to N-[(3S,4R,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]butanamide [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 4GVI_A | 3.27e-18 | 117 | 394 | 18 | 292 | Crystalstructure of mutant (D248N) Salmonella typhimurium family 3 glycoside hydrolase (NagZ) in complex with GlcNAc-1,6-anhMurNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],4GVI_B Crystal structure of mutant (D248N) Salmonella typhimurium family 3 glycoside hydrolase (NagZ) in complex with GlcNAc-1,6-anhMurNAc [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L7N6B0 | 1.55e-19 | 21 | 419 | 7 | 380 | Beta-hexosaminidase LpqI OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lpqI PE=1 SV=1 |
| Q3J953 | 1.72e-19 | 103 | 395 | 6 | 297 | Beta-hexosaminidase OS=Nitrosococcus oceani (strain ATCC 19707 / BCRC 17464 / JCM 30415 / NCIMB 11848 / C-107) OX=323261 GN=nagZ PE=3 SV=1 |
| A0A0H3M1P5 | 3.84e-19 | 21 | 419 | 7 | 380 | Beta-hexosaminidase LpqI OS=Mycobacterium bovis (strain BCG / Pasteur 1173P2) OX=410289 GN=lpqI PE=3 SV=1 |
| B2I6G9 | 4.00e-19 | 109 | 403 | 7 | 299 | Beta-hexosaminidase OS=Xylella fastidiosa (strain M23) OX=405441 GN=nagZ PE=3 SV=1 |
| Q87BR5 | 4.00e-19 | 109 | 403 | 7 | 299 | Beta-hexosaminidase OS=Xylella fastidiosa (strain Temecula1 / ATCC 700964) OX=183190 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
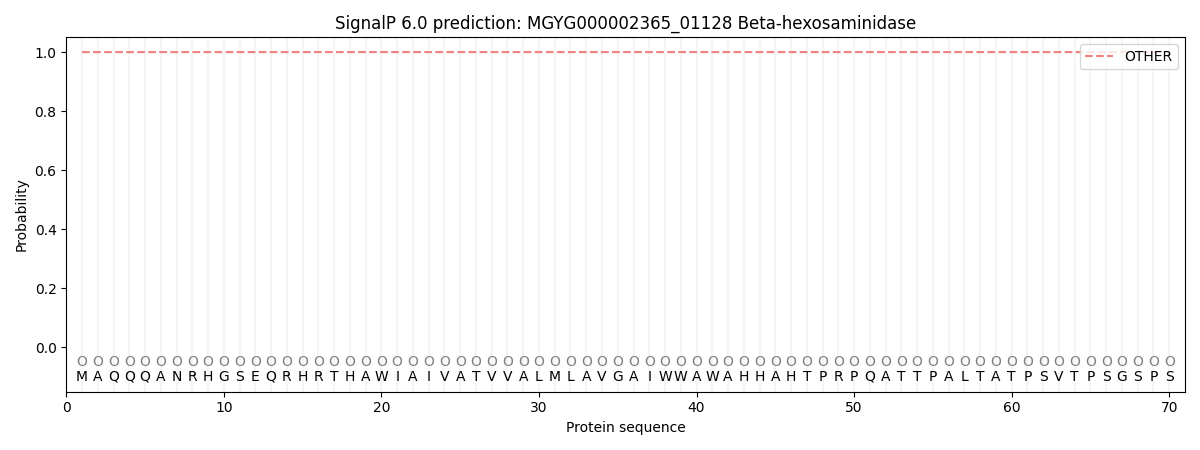
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999990 | 0.000032 | 0.000000 | 0.000000 | 0.000000 | 0.000005 |