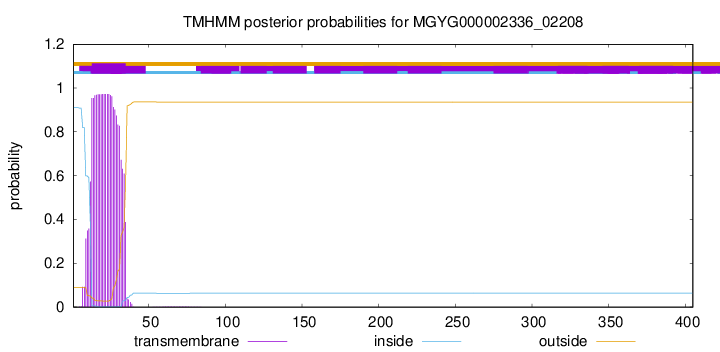You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002336_02208
You are here: Home > Sequence: MGYG000002336_02208
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Megasphaera sp000417505 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Megasphaeraceae; Megasphaera; Megasphaera sp000417505 | |||||||||||
| CAZyme ID | MGYG000002336_02208 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5461; End: 6678 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 101 | 224 | 3.9e-16 | 0.9140625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| sd00006 | TPR | 7.20e-20 | 255 | 351 | 1 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| sd00006 | TPR | 1.20e-17 | 289 | 383 | 1 | 95 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| pfam01832 | Glucosaminidase | 1.73e-12 | 101 | 167 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| COG0457 | TPR | 1.52e-11 | 245 | 397 | 85 | 242 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
| sd00006 | TPR | 2.67e-11 | 252 | 317 | 32 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVO26228.1 | 8.95e-259 | 1 | 405 | 1 | 405 |
| CCC73641.1 | 2.47e-258 | 1 | 405 | 1 | 404 |
| AVO73547.1 | 2.47e-258 | 1 | 405 | 1 | 404 |
| ALG42989.1 | 4.09e-257 | 1 | 405 | 1 | 404 |
| BAL84275.1 | 5.88e-116 | 39 | 402 | 50 | 415 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 5.98e-19 | 80 | 214 | 52 | 179 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
| Q6ZXV5 | 7.25e-06 | 215 | 383 | 514 | 695 | Protein O-mannosyl-transferase TMTC3 OS=Homo sapiens OX=9606 GN=TMTC3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000980 | 0.997876 | 0.000326 | 0.000387 | 0.000212 | 0.000181 |