You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002171_01131
You are here: Home > Sequence: MGYG000002171_01131
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp002493165 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp002493165 | |||||||||||
| CAZyme ID | MGYG000002171_01131 | |||||||||||
| CAZy Family | GH29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24792; End: 26180 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH29 | 23 | 343 | 1.2e-107 | 0.8872832369942196 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01120 | Alpha_L_fucos | 1.56e-112 | 26 | 339 | 19 | 333 | Alpha-L-fucosidase. |
| smart00812 | Alpha_L_fucos | 5.24e-108 | 26 | 410 | 20 | 384 | Alpha-L-fucosidase. O-Glycosyl hydrolases (EC 3.2.1.-) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families. This classification is available on the CAZy (CArbohydrate-Active EnZymes) web site. Because the fold of proteins is better conserved than their sequences, some of the families can be grouped in 'clans'. Family 29 encompasses alpha-L-fucosidases, which is a lysosomal enzyme responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. Deficiency of alpha-L-fucosidase results in the lysosomal storage disease fucosidosis. |
| COG3669 | AfuC | 7.60e-47 | 42 | 447 | 19 | 428 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
| pfam16757 | Fucosidase_C | 2.62e-05 | 382 | 458 | 7 | 90 | Alpha-L-fucosidase C-terminal domain. The C-terminal domain of Structure 1hl8 is constructed of eight anti-parallel-strands packed into two-sheets of five and three strands, respectively, forming a two-layer-sandwich containing a Greek key motif. |
| pfam14871 | GHL6 | 8.87e-04 | 85 | 148 | 1 | 62 | Hypothetical glycosyl hydrolase 6. GHL6 is a family of hypothetical glycoside hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRP56365.1 | 5.11e-240 | 23 | 458 | 23 | 458 |
| QUU06879.1 | 5.11e-240 | 23 | 458 | 23 | 458 |
| ASM65083.1 | 5.11e-240 | 23 | 458 | 23 | 458 |
| QQT78390.1 | 5.11e-240 | 23 | 458 | 23 | 458 |
| SCD21406.1 | 1.05e-211 | 23 | 459 | 26 | 462 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6O1J_A | 3.79e-89 | 23 | 329 | 2 | 334 | ChainA, AlfC [Lacticaseibacillus casei],6O1J_B Chain B, AlfC [Lacticaseibacillus casei],6O1J_C Chain C, AlfC [Lacticaseibacillus casei],6O1J_D Chain D, AlfC [Lacticaseibacillus casei],6O1J_E Chain E, AlfC [Lacticaseibacillus casei],6O1J_F Chain F, AlfC [Lacticaseibacillus casei],6O1J_G Chain G, AlfC [Lacticaseibacillus casei],6O1J_H Chain H, AlfC [Lacticaseibacillus casei] |
| 6O18_A | 3.79e-89 | 23 | 329 | 2 | 334 | ChainA, AlfC [Lacticaseibacillus casei],6O18_B Chain B, AlfC [Lacticaseibacillus casei],6O18_C Chain C, AlfC [Lacticaseibacillus casei],6O18_D Chain D, AlfC [Lacticaseibacillus casei],6O1A_A Chain A, AlfC [Lacticaseibacillus casei],6O1A_B Chain B, AlfC [Lacticaseibacillus casei],6O1A_C Chain C, AlfC [Lacticaseibacillus casei],6O1A_D Chain D, AlfC [Lacticaseibacillus casei] |
| 6O1I_A | 2.13e-88 | 23 | 329 | 2 | 334 | ChainA, AlfC [Lacticaseibacillus casei],6O1I_B Chain B, AlfC [Lacticaseibacillus casei],6O1I_C Chain C, AlfC [Lacticaseibacillus casei],6O1I_D Chain D, AlfC [Lacticaseibacillus casei] |
| 6O1C_A | 5.99e-88 | 23 | 329 | 2 | 334 | ChainA, AlfC [Lacticaseibacillus casei],6O1C_B Chain B, AlfC [Lacticaseibacillus casei],6O1C_C Chain C, AlfC [Lacticaseibacillus casei],6O1C_D Chain D, AlfC [Lacticaseibacillus casei],6O1C_E Chain E, AlfC [Lacticaseibacillus casei],6O1C_F Chain F, AlfC [Lacticaseibacillus casei],6O1C_G Chain G, AlfC [Lacticaseibacillus casei],6O1C_H Chain H, AlfC [Lacticaseibacillus casei],6OHE_A Chain A, AlfC [Lacticaseibacillus casei],6OHE_B Chain B, AlfC [Lacticaseibacillus casei],6OHE_C Chain C, AlfC [Lacticaseibacillus casei],6OHE_D Chain D, AlfC [Lacticaseibacillus casei] |
| 6GN6_A | 9.38e-83 | 21 | 423 | 26 | 401 | Alpha-L-fucosidaseisoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_B Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_C Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_D Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_E Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_F Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P17164 | 3.64e-41 | 27 | 452 | 47 | 450 | Tissue alpha-L-fucosidase OS=Rattus norvegicus OX=10116 GN=Fuca1 PE=1 SV=1 |
| Q99LJ1 | 5.68e-40 | 27 | 452 | 37 | 440 | Tissue alpha-L-fucosidase OS=Mus musculus OX=10090 GN=Fuca1 PE=1 SV=1 |
| P10901 | 3.34e-39 | 28 | 353 | 36 | 372 | Alpha-L-fucosidase OS=Dictyostelium discoideum OX=44689 GN=alfA PE=3 SV=1 |
| Q9BTY2 | 9.69e-39 | 23 | 373 | 44 | 399 | Plasma alpha-L-fucosidase OS=Homo sapiens OX=9606 GN=FUCA2 PE=1 SV=2 |
| Q5RFI5 | 1.30e-38 | 23 | 373 | 42 | 397 | Plasma alpha-L-fucosidase OS=Pongo abelii OX=9601 GN=FUCA2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
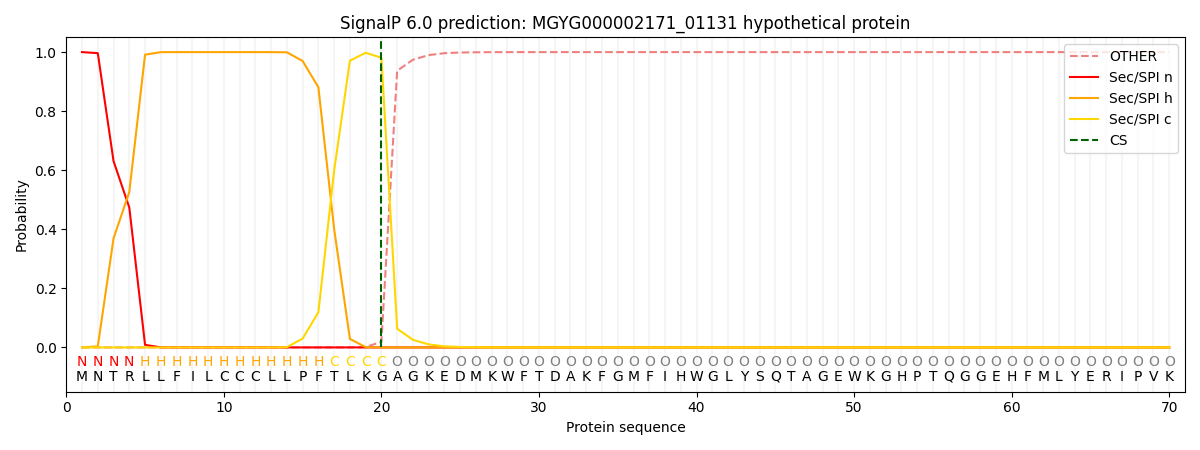
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000188 | 0.999198 | 0.000168 | 0.000146 | 0.000142 | 0.000134 |
