You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001876_00783
You are here: Home > Sequence: MGYG000001876_00783
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acetatifactor sp900760705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; Acetatifactor sp900760705 | |||||||||||
| CAZyme ID | MGYG000001876_00783 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49241; End: 51601 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 73 | 530 | 2.2e-132 | 0.9938900203665988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17996 | CE2_N | 8.70e-05 | 703 | 771 | 24 | 88 | Carbohydrate esterase 2 N-terminal. This is the N-terminal beta-sheet domain with jelly roll topology found in CE2 acetyl-esterase from the bacterium Clostridium thermocellum. This enzyme displays dual activities, it catalyses the deacetylation of plant polysaccharides and also potentiates the activity of its appended cellulase catalytic module through its noncatalytic cellulose binding function. This N-terminal jelly-roll domain appears to extend the substrate/cellulose binding cleft of the catalytic domain in C.thermocellum. |
| pfam13229 | Beta_helix | 0.001 | 156 | 290 | 24 | 139 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| COG3866 | PelB | 0.007 | 172 | 473 | 55 | 327 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam13229 | Beta_helix | 0.010 | 220 | 401 | 1 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDS99205.1 | 1.23e-119 | 69 | 531 | 24 | 510 |
| QGQ95108.1 | 1.20e-117 | 75 | 622 | 39 | 605 |
| QUH27719.1 | 6.65e-117 | 69 | 588 | 30 | 565 |
| QNK55089.1 | 4.54e-116 | 75 | 529 | 256 | 735 |
| BBH21553.1 | 2.00e-114 | 75 | 533 | 43 | 530 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 5.69e-62 | 75 | 530 | 11 | 576 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 1.24e-42 | 75 | 530 | 20 | 596 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 2.35e-40 | 75 | 530 | 16 | 608 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 1.35e-32 | 75 | 403 | 247 | 638 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 7.54e-32 | 75 | 403 | 247 | 638 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR5 | 9.76e-13 | 537 | 780 | 699 | 949 | A type blood alpha-D-galactosamine galactosaminidase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
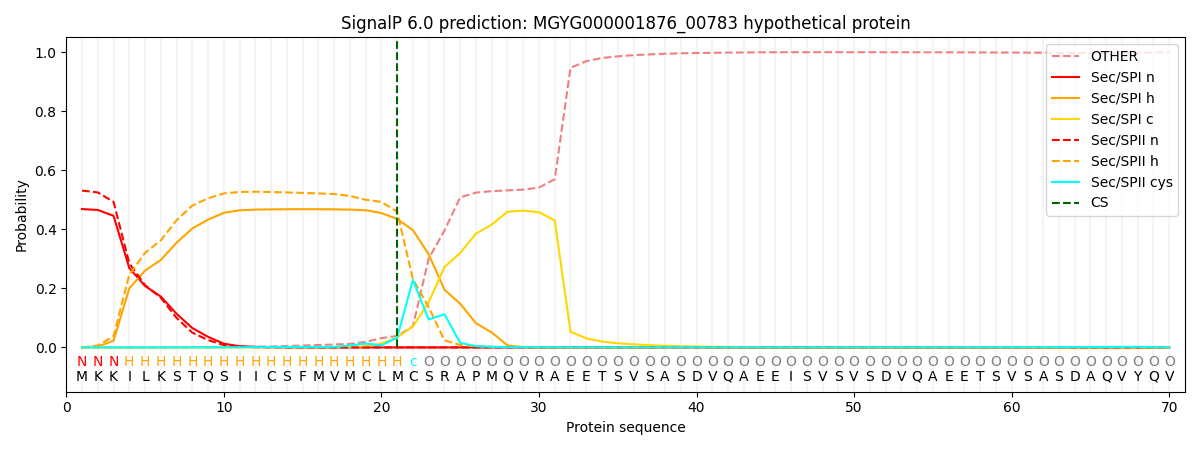
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001335 | 0.456571 | 0.540935 | 0.000541 | 0.000346 | 0.000258 |
