You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001807_00651
You are here: Home > Sequence: MGYG000001807_00651
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000001807_00651 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7768; End: 10011 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 507 | 719 | 4.8e-16 | 0.8722466960352423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00930 | DPPIV_N | 6.05e-57 | 123 | 452 | 18 | 346 | Dipeptidyl peptidase IV (DPP IV) N-terminal region. This family is an alignment of the region to the N-terminal side of the active site. The Prosite motif does not correspond to this Pfam entry. |
| COG1506 | DAP2 | 2.14e-41 | 158 | 717 | 57 | 594 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam00326 | Peptidase_S9 | 5.08e-40 | 542 | 736 | 3 | 210 | Prolyl oligopeptidase family. |
| COG0412 | DLH | 1.30e-05 | 549 | 730 | 50 | 215 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam12146 | Hydrolase_4 | 7.80e-04 | 667 | 721 | 182 | 235 | Serine aminopeptidase, S33. This domain is found in bacteria and eukaryotes and is approximately 110 amino acids in length. It is found in association with pfam00561. The majority of the members in this family carry the exopeptidase active-site residues of Ser-122, Asp-239 and His-269 as in UniProtKB:Q7ZWC2. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AJY87077.1 | 3.72e-212 | 72 | 745 | 270 | 950 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5YP1_A | 2.03e-56 | 165 | 736 | 158 | 743 | Crystalstructure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP1_B Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP1_C Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP1_D Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP2_A Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP2_B Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP3_A Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP3_B Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP3_C Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP3_D Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana [Pseudoxanthomonas mexicana],5YP4_A Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP4_B Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP4_C Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana],5YP4_D Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 [Pseudoxanthomonas mexicana] |
| 2D5L_A | 2.02e-55 | 160 | 722 | 120 | 690 | CrystalStructure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis [Porphyromonas gingivalis W83],2EEP_A Prolyl Tripeptidyl Aminopeptidase Complexed with an Inhibitor [Porphyromonas gingivalis W83] |
| 2ECF_A | 2.32e-55 | 165 | 719 | 156 | 719 | CrystalStructure of Dipeptidyl Aminopeptidase IV from Stenotrophomonas maltophilia [Stenotrophomonas maltophilia] |
| 2Z3W_A | 3.76e-55 | 160 | 722 | 120 | 690 | ChainA, Dipeptidyl aminopeptidase IV [Porphyromonas gingivalis W83],2Z3Z_A Chain A, Dipeptidyl aminopeptidase IV [Porphyromonas gingivalis W83] |
| 2DCM_A | 5.14e-55 | 160 | 722 | 120 | 690 | ChainA, dipeptidyl aminopeptidase IV, putative [Porphyromonas gingivalis W83] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6F3I7 | 1.11e-55 | 165 | 736 | 158 | 743 | Dipeptidyl aminopeptidase 4 OS=Pseudoxanthomonas mexicana OX=128785 GN=dap4 PE=1 SV=1 |
| Q7MUW6 | 1.55e-54 | 160 | 722 | 146 | 716 | Prolyl tripeptidyl peptidase OS=Porphyromonas gingivalis (strain ATCC BAA-308 / W83) OX=242619 GN=ptpA PE=1 SV=1 |
| B2RJX3 | 1.36e-53 | 160 | 722 | 146 | 716 | Prolyl tripeptidyl peptidase OS=Porphyromonas gingivalis (strain ATCC 33277 / DSM 20709 / CIP 103683 / JCM 12257 / NCTC 11834 / 2561) OX=431947 GN=ptpA PE=3 SV=1 |
| P97321 | 1.74e-32 | 46 | 736 | 54 | 753 | Prolyl endopeptidase FAP OS=Mus musculus OX=10090 GN=Fap PE=1 SV=1 |
| Q6V1X1 | 2.32e-31 | 324 | 740 | 433 | 887 | Dipeptidyl peptidase 8 OS=Homo sapiens OX=9606 GN=DPP8 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
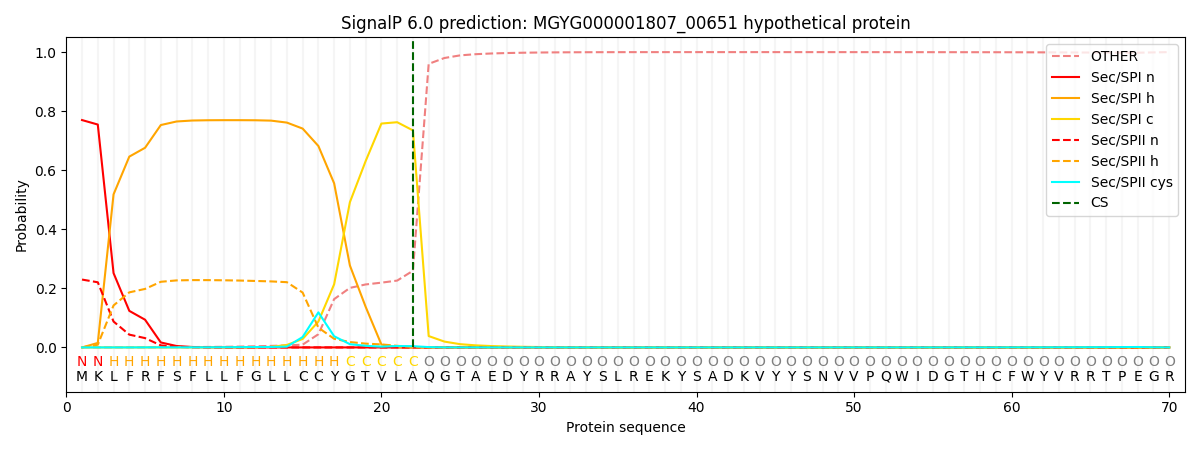
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001441 | 0.761241 | 0.236453 | 0.000332 | 0.000261 | 0.000253 |
