You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001780_00111
You are here: Home > Sequence: MGYG000001780_00111
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000001780_00111 | |||||||||||
| CAZy Family | PL6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 117751; End: 119145 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL6 | 39 | 378 | 2.9e-106 | 0.9538904899135446 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14251 | PL-6 | 2.81e-154 | 37 | 399 | 11 | 364 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam14592 | Chondroitinas_B | 5.50e-90 | 39 | 399 | 13 | 378 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
| pfam13229 | Beta_helix | 6.63e-06 | 224 | 317 | 32 | 123 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ58476.1 | 0.0 | 1 | 464 | 1 | 464 |
| QDO71143.1 | 3.27e-317 | 1 | 464 | 2 | 465 |
| QUT90411.1 | 1.35e-315 | 1 | 464 | 1 | 464 |
| QDO71096.1 | 1.23e-150 | 18 | 424 | 21 | 433 |
| AHY60627.1 | 2.09e-75 | 39 | 450 | 31 | 439 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6QPS_A | 1.54e-307 | 19 | 464 | 22 | 467 | Structuralcharacterization of a mannuronic acid specific polysaccharide family 6 lyase enzyme from human gut microbiota [Bacteroides cellulosilyticus],6QPS_B Structural characterization of a mannuronic acid specific polysaccharide family 6 lyase enzyme from human gut microbiota [Bacteroides cellulosilyticus] |
| 5GKD_A | 1.84e-63 | 43 | 399 | 21 | 369 | Structureof PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_B Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_C Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_D Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis] |
| 5GKQ_A | 1.31e-62 | 43 | 399 | 21 | 369 | Structureof PL6 family alginate lyase AlyGC mutant-R241A [Paraglaciecola chathamensis S18K6],5GKQ_B Structure of PL6 family alginate lyase AlyGC mutant-R241A [Paraglaciecola chathamensis S18K6] |
| 7DMK_A | 1.72e-61 | 43 | 422 | 30 | 391 | ChainA, BcAlyPL6 [Bacteroides clarus],7DMK_B Chain B, BcAlyPL6 [Bacteroides clarus],7DMK_C Chain C, BcAlyPL6 [Bacteroides clarus],7DMK_D Chain D, BcAlyPL6 [Bacteroides clarus] |
| 7O84_A | 8.26e-61 | 35 | 436 | 27 | 405 | ChainA, Alginate lyase [Pseudopedobacter saltans DSM 12145],7O84_B Chain B, Alginate lyase [Pseudopedobacter saltans DSM 12145] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q06365 | 1.20e-53 | 43 | 389 | 3 | 334 | Alginate lyase OS=Pseudomonas sp. (strain OS-ALG-9) OX=86038 GN=aly PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
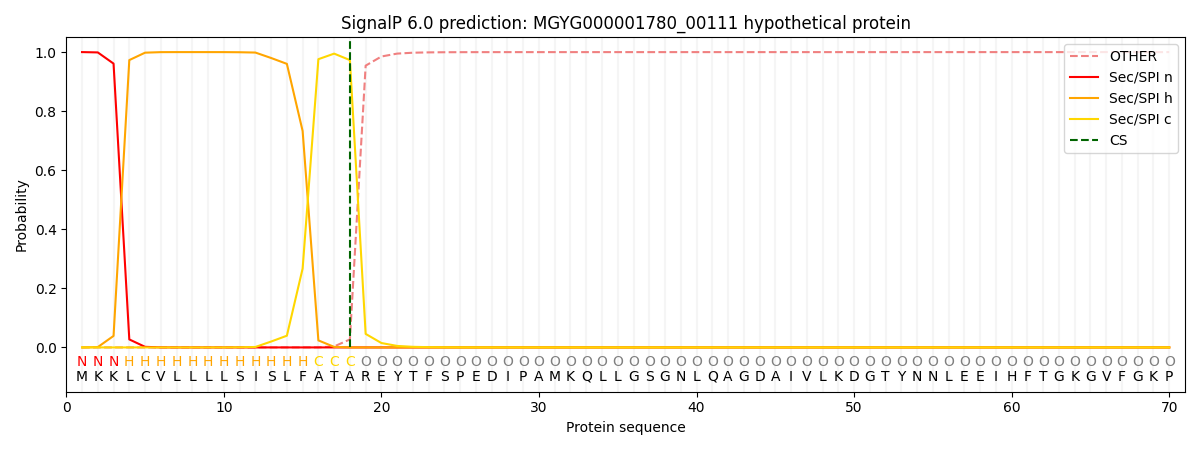
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000306 | 0.998912 | 0.000275 | 0.000155 | 0.000159 | 0.000149 |
