You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001748_01909
You are here: Home > Sequence: MGYG000001748_01909
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
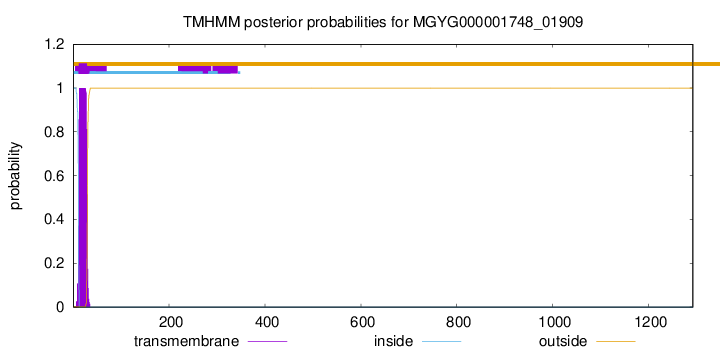
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900762665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900762665 | |||||||||||
| CAZyme ID | MGYG000001748_01909 | |||||||||||
| CAZy Family | GH120 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47903; End: 51781 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 950 | 1292 | 3.3e-47 | 0.4487534626038781 |
| CBM32 | 626 | 739 | 2.9e-27 | 0.9274193548387096 |
| GH120 | 323 | 416 | 2.4e-17 | 0.989010989010989 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14498 | Glyco_hyd_65N_2 | 2.13e-21 | 956 | 1185 | 8 | 224 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
| pfam00754 | F5_F8_type_C | 8.84e-20 | 624 | 737 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam05345 | He_PIG | 1.11e-08 | 751 | 836 | 1 | 94 | Putative Ig domain. This alignment represents the conserved core region of ~90 residue repeat found in several haemagglutinins and other cell surface proteins. Sequence similarities to (pfam02494) and (pfam00801) suggest an Ig-like fold (personal obs:C. Yeats). So this family may be similar in function to the (pfam02639) and (pfam02638) domains. This domain is also found in the WisP family of proteins of Tropheryma whipplei. |
| cd00063 | FN3 | 2.28e-07 | 855 | 921 | 8 | 81 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| cd00057 | FA58C | 2.89e-07 | 624 | 742 | 13 | 143 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWS40336.1 | 0.0 | 29 | 1291 | 31 | 1346 |
| BCB78141.1 | 6.75e-228 | 37 | 742 | 42 | 742 |
| ASY32077.1 | 1.48e-220 | 46 | 758 | 53 | 761 |
| BBE20411.1 | 4.89e-100 | 963 | 1292 | 1 | 326 |
| ANI89979.1 | 4.10e-97 | 946 | 1292 | 29 | 371 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VST_A | 6.56e-79 | 40 | 527 | 3 | 507 | Thecomplex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_B The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_C The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_D The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_A The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_B The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_C The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_D The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_A The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_B The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_C The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_D The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 2RVA_A | 1.09e-28 | 618 | 742 | 7 | 137 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZZ5_A | 1.12e-28 | 618 | 742 | 8 | 138 | X-raycrystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ5_B X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_A X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_B X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 2RV9_A | 2.67e-28 | 618 | 742 | 7 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 2.75e-28 | 618 | 742 | 8 | 137 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 2.31e-06 | 828 | 944 | 541 | 654 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P94576 | 9.35e-06 | 9 | 112 | 2 | 107 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
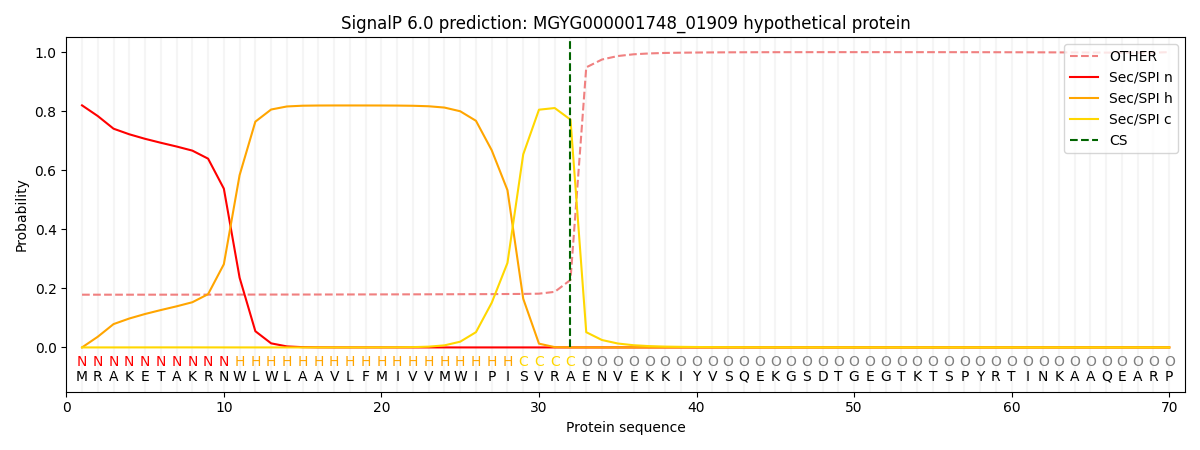
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.194292 | 0.800921 | 0.003288 | 0.000600 | 0.000392 | 0.000477 |