You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001748_00130
You are here: Home > Sequence: MGYG000001748_00130
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
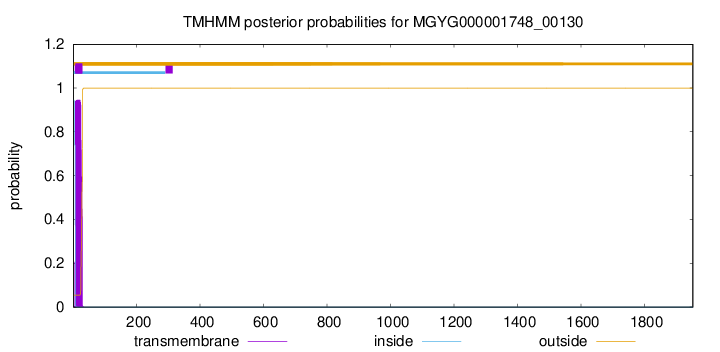
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900762665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900762665 | |||||||||||
| CAZyme ID | MGYG000001748_00130 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 157476; End: 163337 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 39 | 604 | 4.9e-124 | 0.9905123339658444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| sd00036 | LRR_3 | 1.07e-13 | 1840 | 1915 | 14 | 102 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| pfam13306 | LRR_5 | 2.24e-12 | 1861 | 1915 | 1 | 53 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
| pfam13306 | LRR_5 | 6.62e-12 | 1843 | 1915 | 37 | 120 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
| pfam13306 | LRR_5 | 7.08e-12 | 1838 | 1915 | 9 | 75 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
| sd00036 | LRR_3 | 9.37e-12 | 1843 | 1915 | 63 | 125 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUX40294.1 | 5.53e-163 | 41 | 775 | 19 | 730 |
| AEY67284.1 | 5.08e-162 | 2 | 788 | 8 | 772 |
| ADI13073.1 | 5.44e-162 | 10 | 767 | 14 | 761 |
| AUX31799.1 | 1.10e-160 | 41 | 775 | 97 | 808 |
| ACL76007.1 | 4.70e-160 | 2 | 788 | 8 | 772 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 5.74e-43 | 30 | 628 | 12 | 601 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
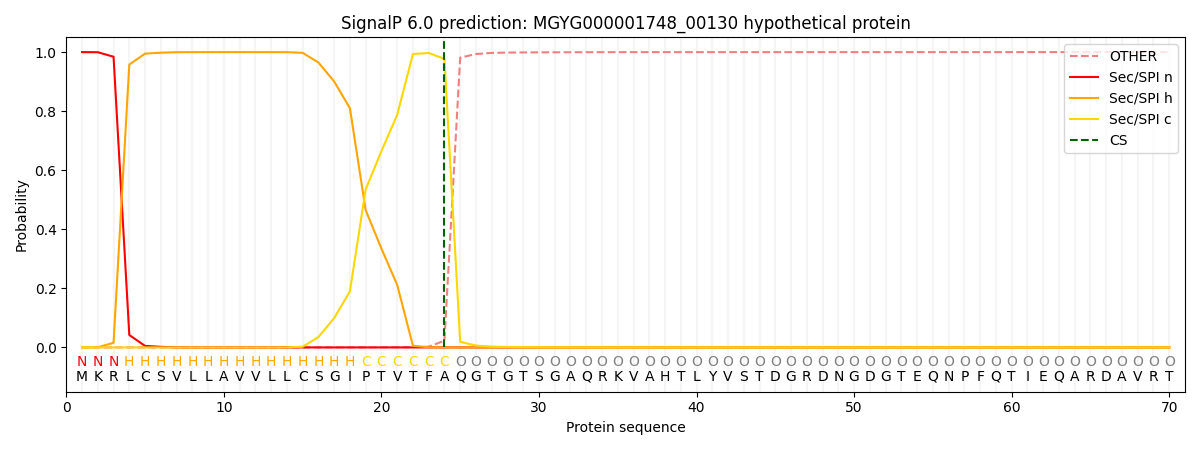
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000230 | 0.998937 | 0.000290 | 0.000179 | 0.000155 | 0.000135 |