You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001561_00978
You are here: Home > Sequence: MGYG000001561_00978
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
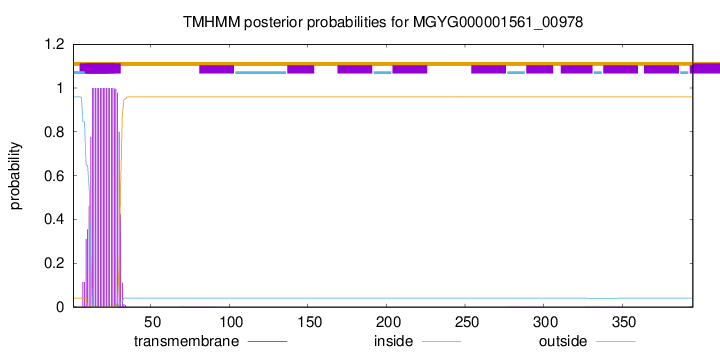
TMHMM annotations
Basic Information help
| Species | Caecibacter massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Megasphaeraceae; Caecibacter; Caecibacter massiliensis | |||||||||||
| CAZyme ID | MGYG000001561_00978 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1029617; End: 1030804 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 125 | 347 | 2.1e-48 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 6.65e-72 | 63 | 388 | 1 | 314 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 4.30e-68 | 68 | 384 | 5 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 2.93e-55 | 74 | 350 | 5 | 282 | beta-hexosaminidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXB83100.1 | 2.71e-280 | 1 | 395 | 1 | 395 |
| CCC72588.1 | 8.53e-162 | 1 | 387 | 1 | 379 |
| AVO26856.1 | 8.53e-162 | 1 | 387 | 1 | 379 |
| AVO74039.1 | 8.53e-162 | 1 | 387 | 1 | 379 |
| ALG41456.1 | 1.40e-160 | 1 | 387 | 1 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 9.06e-57 | 61 | 386 | 9 | 337 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 4ZM6_A | 3.42e-45 | 69 | 386 | 13 | 337 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
| 3TEV_A | 6.25e-44 | 68 | 384 | 17 | 329 | Thecrystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1],3TEV_B The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1] |
| 5BU9_A | 7.76e-40 | 63 | 386 | 5 | 337 | Crystalstructure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333],5BU9_B Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333] |
| 7VI6_A | 1.25e-37 | 74 | 374 | 5 | 306 | ChainA, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI6_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_A Chain A, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835],7VI7_B Chain B, Beta-N-acetylhexosaminidase [Akkermansia muciniphila ATCC BAA-835] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48823 | 2.66e-39 | 80 | 386 | 49 | 380 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| P58067 | 1.64e-38 | 73 | 350 | 4 | 281 | Beta-hexosaminidase OS=Escherichia coli O157:H7 OX=83334 GN=nagZ PE=3 SV=1 |
| B5YVX6 | 1.64e-38 | 73 | 350 | 4 | 281 | Beta-hexosaminidase OS=Escherichia coli O157:H7 (strain EC4115 / EHEC) OX=444450 GN=nagZ PE=3 SV=1 |
| B2VDM3 | 1.71e-38 | 73 | 350 | 4 | 281 | Beta-hexosaminidase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=nagZ PE=3 SV=1 |
| Q8FIN2 | 2.29e-38 | 73 | 350 | 4 | 281 | Beta-hexosaminidase OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
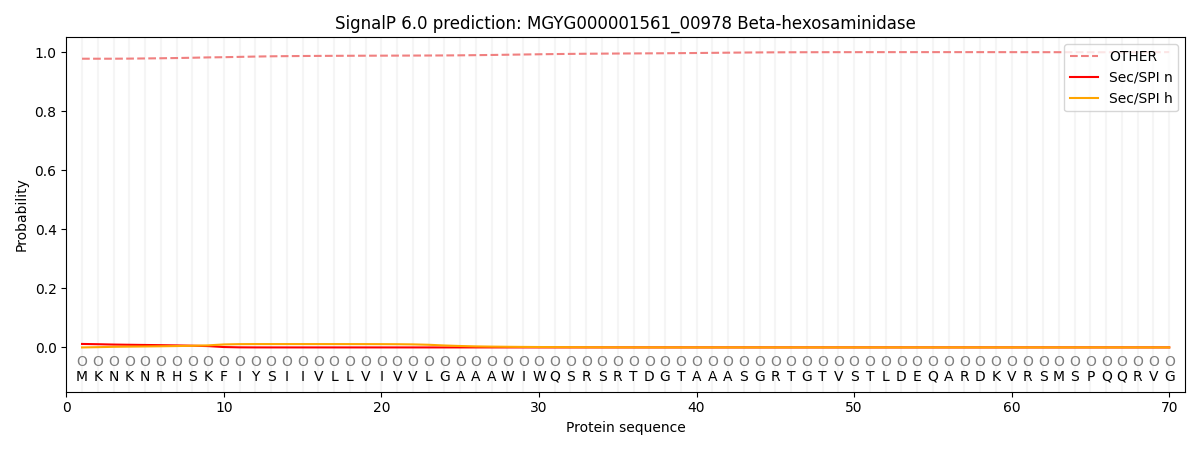
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.978137 | 0.010760 | 0.001919 | 0.000114 | 0.000061 | 0.009015 |