You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001548_03829
You are here: Home > Sequence: MGYG000001548_03829
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A tuaregi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A tuaregi | |||||||||||
| CAZyme ID | MGYG000001548_03829 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4200850; End: 4205163 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 435 | 732 | 6.8e-66 | 0.8943894389438944 |
| CBM23 | 1100 | 1259 | 1.4e-37 | 0.9876543209876543 |
| CBM54 | 198 | 310 | 7.9e-32 | 0.9824561403508771 |
| CBM59 | 1286 | 1432 | 4.3e-27 | 0.993103448275862 |
| CBM27 | 913 | 1087 | 6.3e-19 | 0.9880952380952381 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 4.13e-54 | 435 | 740 | 1 | 299 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 8.57e-21 | 513 | 745 | 93 | 333 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| NF033190 | inl_like_NEAT_1 | 2.88e-11 | 42 | 196 | 588 | 745 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 1.49e-08 | 155 | 195 | 1 | 42 | S-layer homology domain. |
| pfam09212 | CBM27 | 8.01e-07 | 909 | 1088 | 7 | 171 | Carbohydrate binding module 27. Members of this family are carbohydrate binding modules that bind to beta-1, 4-manno-oligosaccharides, carob galactomannan, and konjac glucomannan, but not to cellulose (insoluble and soluble) or soluble birchwood xylan. They adopt a beta sandwich structure comprising 13 beta strands with a single, small alpha-helix and a single metal atom. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFC29293.1 | 0.0 | 42 | 1432 | 41 | 1425 |
| AEI40679.1 | 0.0 | 42 | 1432 | 41 | 1419 |
| QMV42451.1 | 0.0 | 42 | 1435 | 44 | 1399 |
| ANY66447.1 | 0.0 | 2 | 1436 | 3 | 1404 |
| ADL67722.1 | 0.0 | 43 | 1432 | 53 | 1408 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 1.46e-141 | 434 | 818 | 51 | 441 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 2BVT_A | 3.76e-126 | 434 | 908 | 8 | 469 | Thestructure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVT_B The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVY_A The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi [Cellulomonas fimi] |
| 2X2Y_A | 2.68e-124 | 434 | 908 | 8 | 469 | Cellulomonasfimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi],2X2Y_B Cellulomonas fimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi] |
| 3TP4_A | 1.14e-113 | 434 | 908 | 8 | 469 | CrystalStructure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct],3TP4_B Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct] |
| 1J9Y_A | 1.75e-89 | 434 | 796 | 11 | 373 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49424 | 3.08e-88 | 434 | 796 | 49 | 411 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| A1A278 | 8.11e-82 | 433 | 1194 | 41 | 813 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| C6CRV0 | 5.94e-18 | 44 | 202 | 1291 | 1458 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P16699 | 3.02e-17 | 429 | 753 | 29 | 328 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| P38537 | 4.12e-16 | 44 | 210 | 42 | 213 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
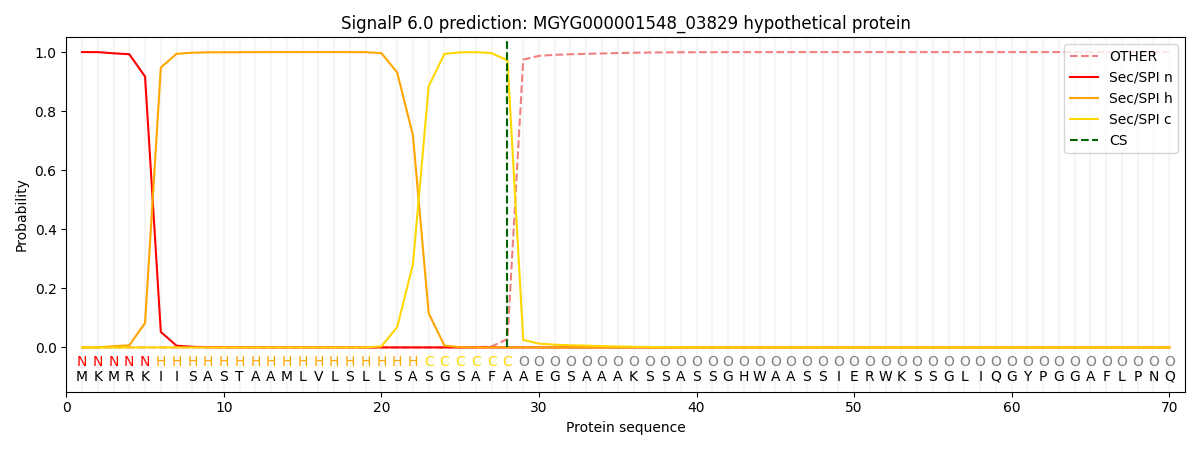
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000275 | 0.998997 | 0.000201 | 0.000194 | 0.000170 | 0.000153 |

