You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_03522
You are here: Home > Sequence: MGYG000001537_03522
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_03522 | |||||||||||
| CAZy Family | GH74 | |||||||||||
| CAZyme Description | Xyloglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 860895; End: 863639 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM2 | 812 | 911 | 6.4e-34 | 0.9900990099009901 |
| GH74 | 92 | 208 | 5.3e-26 | 0.48497854077253216 |
| GH74 | 578 | 697 | 1.8e-20 | 0.5107296137339056 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00553 | CBM_2 | 1.57e-33 | 812 | 911 | 1 | 101 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart00637 | CBD_II | 4.50e-27 | 818 | 910 | 1 | 92 | CBD_II domain. |
| COG5297 | CelA1 | 3.55e-07 | 819 | 890 | 472 | 540 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| pfam15902 | Sortilin-Vps10 | 3.22e-05 | 251 | 368 | 1 | 110 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| smart00602 | VPS10 | 2.58e-04 | 132 | 217 | 55 | 135 | VPS10 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHT55758.1 | 0.0 | 15 | 911 | 24 | 928 |
| QGQ19309.1 | 0.0 | 1 | 911 | 1 | 926 |
| AEE46043.1 | 0.0 | 31 | 914 | 40 | 918 |
| VEH31406.1 | 0.0 | 31 | 914 | 40 | 918 |
| AEI13625.1 | 0.0 | 17 | 914 | 21 | 920 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6P2O_A | 0.0 | 41 | 764 | 3 | 727 | Crystalstructure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG [Streptomyces rapamycinicus NRRL 5491],6P2O_B Crystal structure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG [Streptomyces rapamycinicus NRRL 5491] |
| 5JWZ_A | 2.58e-317 | 41 | 762 | 3 | 723 | Structureof a Putative Xyloglucanase from the Cellulolytic Bacteria Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],5JWZ_B Structure of a Putative Xyloglucanase from the Cellulolytic Bacteria Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 5FKR_A | 2.70e-311 | 41 | 762 | 5 | 729 | Unravelingthe first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase [Cellvibrio japonicus Ueda107],5FKS_A Unraveling the first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase [Cellvibrio japonicus Ueda107] |
| 5FKQ_A | 3.33e-310 | 36 | 762 | 2 | 731 | Unravelingthe first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase [Cellvibrio japonicus Ueda107],5FKQ_B Unraveling the first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase [Cellvibrio japonicus Ueda107] |
| 5FKT_A | 3.33e-310 | 36 | 762 | 2 | 731 | Unravelingthe first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase [Cellvibrio japonicus Ueda107],5FKT_B Unraveling the first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase [Cellvibrio japonicus Ueda107] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3MUH7 | 1.91e-288 | 16 | 765 | 11 | 768 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
| A3DFA0 | 8.25e-242 | 41 | 768 | 38 | 763 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xghA PE=3 SV=1 |
| Q70DK5 | 3.30e-241 | 41 | 768 | 38 | 763 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus OX=1515 GN=xghA PE=1 SV=1 |
| Q7Z9M8 | 4.63e-179 | 37 | 654 | 17 | 642 | Xyloglucanase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cel74a PE=1 SV=1 |
| Q5BD38 | 5.66e-104 | 35 | 763 | 26 | 808 | Oligoxyloglucan-reducing end-specific xyloglucanase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgcA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
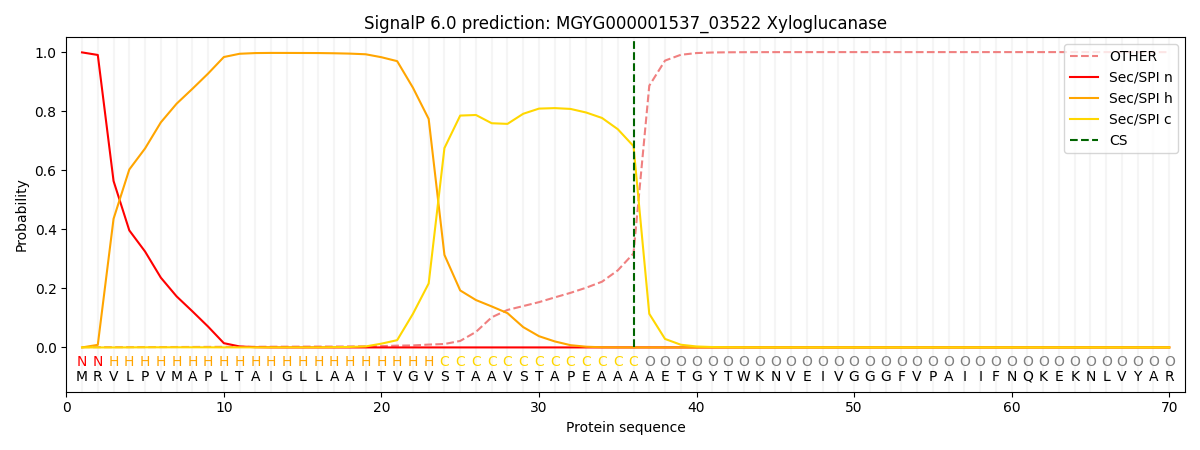
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001833 | 0.997168 | 0.000287 | 0.000286 | 0.000210 | 0.000203 |
