You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_00983
You are here: Home > Sequence: MGYG000001537_00983
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
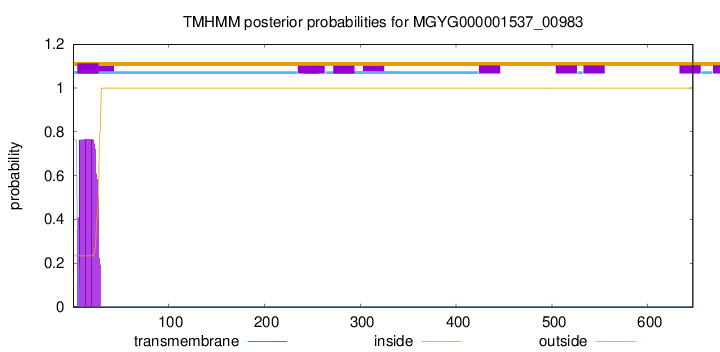
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_00983 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 266564; End: 268507 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 72 | 363 | 2.6e-99 | 0.986159169550173 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 1.20e-51 | 50 | 382 | 6 | 301 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 4.80e-17 | 48 | 366 | 1 | 270 | Cellulase (glycosyl hydrolase family 5). |
| pfam17963 | Big_9 | 1.74e-05 | 408 | 492 | 14 | 87 | Bacterial Ig domain. This entry represents a wide variety of bacterial Ig domains. |
| NF012211 | tand_rpt_95 | 0.009 | 461 | 509 | 39 | 86 | tandem-95 repeat. This 95-amino acid repeat occurs in tandem in proteins that may be several thousand amino acids long. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SBT52970.1 | 1.43e-222 | 40 | 644 | 35 | 656 |
| SCG46694.1 | 1.20e-221 | 40 | 644 | 36 | 657 |
| ACZ31816.1 | 8.58e-221 | 37 | 644 | 50 | 674 |
| BCJ58063.1 | 1.13e-220 | 43 | 644 | 3 | 621 |
| AVT32268.1 | 9.18e-218 | 40 | 644 | 42 | 664 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QP0_A | 8.40e-103 | 37 | 400 | 1 | 357 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 3PZ9_A | 3.76e-98 | 50 | 396 | 22 | 380 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 5.87e-95 | 50 | 396 | 8 | 366 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 3ZIZ_A | 5.76e-67 | 44 | 392 | 19 | 346 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
| 1QNO_A | 8.37e-64 | 37 | 386 | 1 | 329 | ChainA, ENDO-1,4-B-D-MANNANASE [Trichoderma reesei],1QNP_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNQ_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNR_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNS_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5AR04 | 4.43e-67 | 34 | 386 | 84 | 416 | Probable mannan endo-1,4-beta-mannosidase F OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manF PE=3 SV=2 |
| B2B3C0 | 2.45e-66 | 44 | 392 | 33 | 360 | Mannan endo-1,4-beta-mannosidase A OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=Pa_6_490 PE=1 SV=1 |
| Q0CUG5 | 3.32e-66 | 36 | 383 | 43 | 370 | Probable mannan endo-1,4-beta-mannosidase A-2 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=manA-2 PE=3 SV=2 |
| B0Y9E7 | 3.73e-66 | 37 | 383 | 94 | 421 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
| Q4WBS1 | 3.73e-66 | 37 | 383 | 94 | 421 | Mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=manF PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
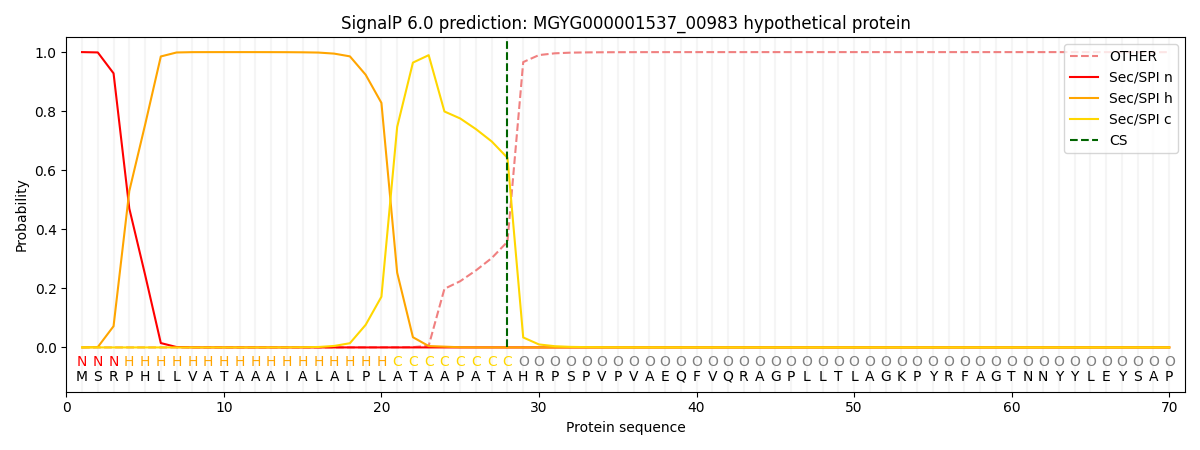
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000479 | 0.998478 | 0.000255 | 0.000318 | 0.000243 | 0.000213 |