You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001525_04857
You are here: Home > Sequence: MGYG000001525_04857
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
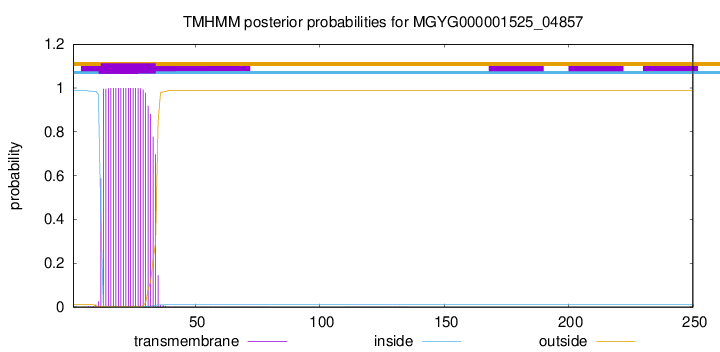
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A rubinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A rubinfantis | |||||||||||
| CAZyme ID | MGYG000001525_04857 | |||||||||||
| CAZy Family | CBM12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1456377; End: 1457129 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd12214 | ChiA1_BD | 7.11e-17 | 40 | 79 | 4 | 43 | chitin-binding domain of Chi A1-like proteins. This group contains proteins related to the chitin binding domain of chitinase A1 (ChiA1) of Bacillus circulans WL-12. Glycosidase ChiA1 hydrolyzes chitin and is comprised of several domains: the C-terminal chitin binding domain, an N-terminal and catalytic domain, and 2 fibronectin type III-like domains. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. Bacillus circulans WL-12 ChiA1 facilitates invasion of fungal cell walls. The ChiAi chitin binding domain is required for the specific recognition of insoluble chitin. although topologically and structurally related, ChiA1 lacks the characteristic aromatic residues of Erwinia chrysanthemi endoglucanase Z (CBD(EGZ)). |
| cd13400 | LT_IagB-like | 1.02e-07 | 130 | 245 | 6 | 109 | Escherichia coli invasion protein IagB and similar proteins. Lytic transglycosylase-like protein, similar to Escherichia coli invasion protein IagB. IagB is encoded within a pathogenicity island in Salmonella enterica and has been shown to degrade polymeric peptidoglycan. IagB-like invasion proteins are implicated in the invasion of eukaryotic host cells by bacteria. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Members of this family resemble the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
| pfam01464 | SLT | 5.17e-04 | 131 | 215 | 14 | 95 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd00254 | LT-like | 6.12e-04 | 131 | 228 | 3 | 90 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| smart00495 | ChtBD3 | 0.002 | 40 | 78 | 6 | 41 | Chitin-binding domain type 3. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAH10650.1 | 7.43e-144 | 1 | 249 | 1 | 251 |
| AUO08603.1 | 1.04e-138 | 1 | 250 | 1 | 250 |
| AJE53774.1 | 4.24e-138 | 1 | 250 | 1 | 250 |
| QOH62288.1 | 4.24e-138 | 1 | 250 | 1 | 250 |
| QNR69760.1 | 4.24e-138 | 1 | 250 | 1 | 250 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W6B_A | 6.16e-98 | 101 | 249 | 32 | 180 | Crystalstructure of catalytic domain of chitinase from Ralstonia sp. A-471 [Ralstonia sp. A-471],3W6B_B Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 [Ralstonia sp. A-471],3W6B_C Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 [Ralstonia sp. A-471],3W6B_D Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 [Ralstonia sp. A-471],3W6C_A Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 in complex with disaccharide [Ralstonia sp. A-471],3W6C_B Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 in complex with disaccharide [Ralstonia sp. A-471],3W6C_C Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 in complex with disaccharide [Ralstonia sp. A-471],3W6C_D Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 in complex with disaccharide [Ralstonia sp. A-471] |
| 3W6D_A | 1.76e-97 | 101 | 249 | 32 | 180 | Crystalstructure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide [Ralstonia sp. A-471],3W6D_B Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide [Ralstonia sp. A-471],3W6D_C Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide [Ralstonia sp. A-471],3W6D_D Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide [Ralstonia sp. A-471] |
| 3W6E_A | 1.76e-97 | 101 | 249 | 32 | 180 | Crystalstructure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) [Ralstonia sp. A-471],3W6E_B Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) [Ralstonia sp. A-471],3W6E_C Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) [Ralstonia sp. A-471],3W6E_D Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) [Ralstonia sp. A-471],3W6F_A Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) in complex with disaccharide [Ralstonia sp. A-471],3W6F_B Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) in complex with disaccharide [Ralstonia sp. A-471],3W6F_C Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) in complex with disaccharide [Ralstonia sp. A-471],3W6F_D Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) in complex with disaccharide [Ralstonia sp. A-471] |
| 1ED7_A | 1.17e-13 | 37 | 78 | 1 | 42 | ChainA, CHITINASE A1 [Niallia circulans] |
| 4ILV_A | 1.40e-12 | 36 | 78 | 203 | 245 | Structureof the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4ILV_B Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 1.13e-10 | 37 | 78 | 655 | 696 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P52320 | 1.03e-09 | 36 | 78 | 412 | 454 | Streptogrisin-C OS=Streptomyces griseus OX=1911 GN=sprC PE=3 SV=1 |
| P27050 | 1.25e-07 | 30 | 82 | 28 | 78 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
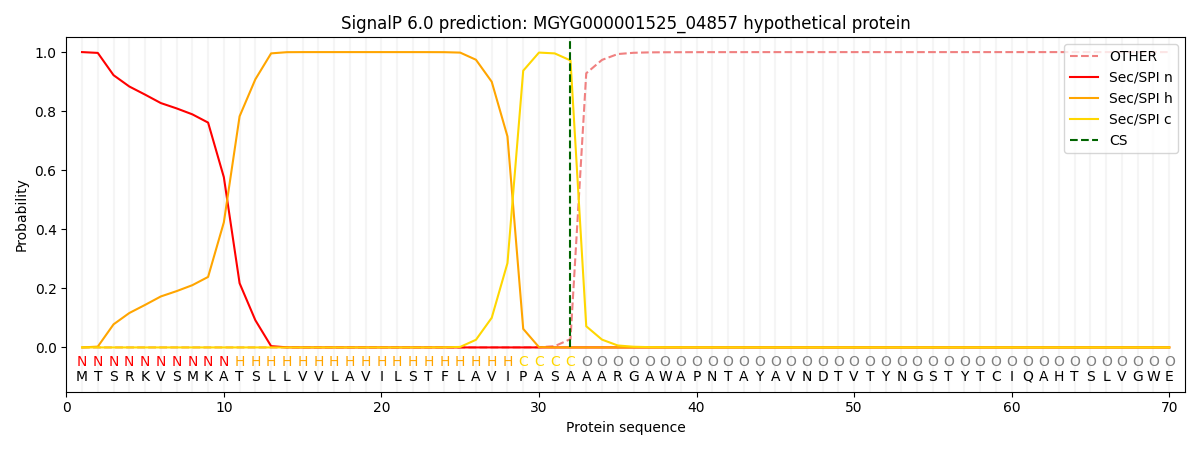
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000235 | 0.999149 | 0.000149 | 0.000178 | 0.000145 | 0.000133 |