You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001525_00542
You are here: Home > Sequence: MGYG000001525_00542
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
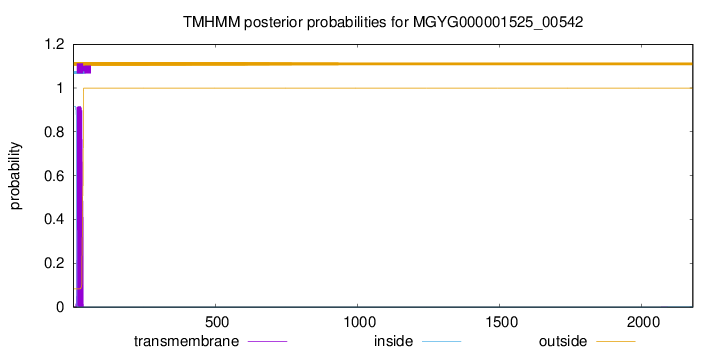
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A rubinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A rubinfantis | |||||||||||
| CAZyme ID | MGYG000001525_00542 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36522; End: 43070 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 154 | 703 | 6.2e-30 | 0.8401162790697675 |
| GH36 | 1001 | 1348 | 2.9e-24 | 0.5406976744186046 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03442 | CBM_X2 | 6.82e-21 | 1675 | 1758 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
| NF033190 | inl_like_NEAT_1 | 1.14e-19 | 2011 | 2169 | 587 | 743 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| cd14791 | GH36 | 4.87e-17 | 382 | 602 | 31 | 261 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| pfam03442 | CBM_X2 | 5.39e-12 | 819 | 906 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
| cd14791 | GH36 | 1.17e-11 | 1238 | 1392 | 31 | 211 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGU28165.1 | 4.73e-189 | 53 | 807 | 32 | 760 |
| QTH40290.1 | 3.80e-118 | 58 | 822 | 37 | 794 |
| QNK59398.1 | 9.16e-111 | 916 | 1667 | 39 | 788 |
| QNA93536.1 | 1.06e-97 | 55 | 829 | 43 | 822 |
| QYM63786.1 | 1.31e-97 | 75 | 829 | 12 | 772 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 1.99e-16 | 2013 | 2170 | 33 | 188 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 2.04e-16 | 2013 | 2170 | 12 | 167 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 3MI6_A | 1.47e-13 | 992 | 1536 | 110 | 696 | ChainA, Alpha-galactosidase [Levilactobacillus brevis ATCC 367],3MI6_B Chain B, Alpha-galactosidase [Levilactobacillus brevis ATCC 367],3MI6_C Chain C, Alpha-galactosidase [Levilactobacillus brevis ATCC 367],3MI6_D Chain D, Alpha-galactosidase [Levilactobacillus brevis ATCC 367] |
| 2YFN_A | 3.76e-12 | 1226 | 1561 | 346 | 705 | galactosidasedomain of alpha-galactosidase-sucrose kinase, AgaSK [[Ruminococcus] gnavus E1],2YFO_A GALACTOSIDASE DOMAIN OF ALPHA-GALACTOSIDASE-SUCROSE KINASE, AGASK, in complex with galactose [[Ruminococcus] gnavus E1] |
| 1K85_A | 4.43e-09 | 1876 | 1947 | 19 | 88 | ChainA, CHITINASE A1 [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6CRV0 | 2.30e-17 | 2014 | 2173 | 1291 | 1454 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| Q06853 | 5.16e-17 | 2005 | 2158 | 523 | 679 | Cell surface glycoprotein 2 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=Cthe_3079 PE=1 SV=1 |
| P38537 | 1.39e-16 | 2014 | 2181 | 42 | 209 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| Q9ZES5 | 1.11e-14 | 2013 | 2170 | 41 | 197 | S-layer protein OS=Bacillus thuringiensis subsp. finitimus OX=29337 GN=ctc PE=1 SV=1 |
| Q06848 | 5.36e-14 | 2005 | 2158 | 283 | 439 | Cellulosome-anchoring protein OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=ancA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
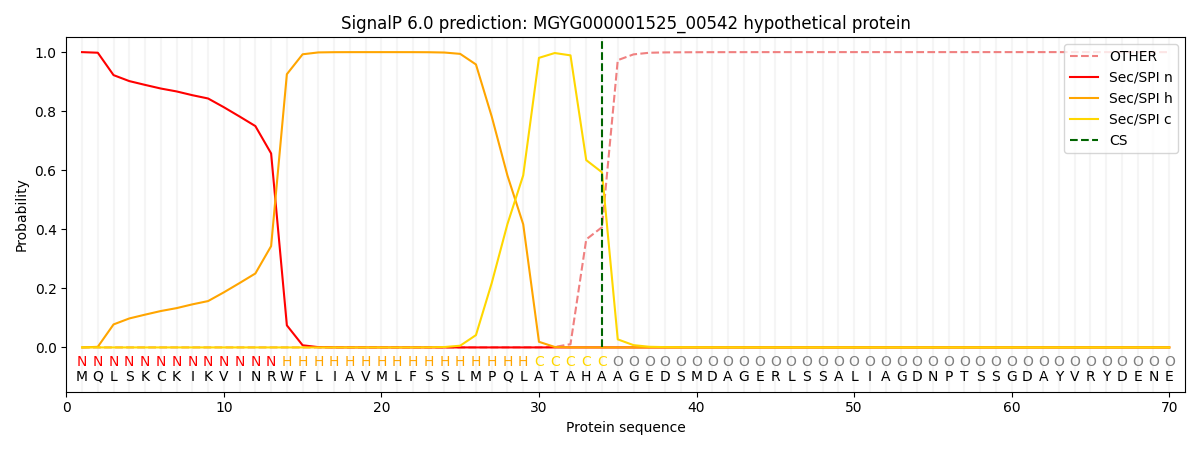
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000556 | 0.998548 | 0.000271 | 0.000224 | 0.000200 | 0.000187 |