You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001514_04851
You are here: Home > Sequence: MGYG000001514_04851
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A ihumii | |||||||||||
| CAZyme ID | MGYG000001514_04851 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3649766; End: 3651556 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 49 | 295 | 5.9e-97 | 0.9881422924901185 |
| CBM3 | 453 | 532 | 7.7e-27 | 0.9886363636363636 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.80e-36 | 48 | 292 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| pfam00942 | CBM_3 | 5.77e-24 | 452 | 531 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 6.17e-23 | 452 | 532 | 1 | 83 | Cellulose binding domain. |
| cd00063 | FN3 | 0.001 | 354 | 421 | 1 | 74 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 0.002 | 354 | 421 | 1 | 74 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMV43160.1 | 3.35e-264 | 28 | 596 | 30 | 676 |
| AWV34670.1 | 1.92e-245 | 13 | 596 | 15 | 525 |
| AIQ75360.1 | 5.81e-242 | 13 | 596 | 15 | 573 |
| CQR54354.1 | 4.82e-241 | 13 | 596 | 14 | 554 |
| AIQ19732.1 | 1.13e-238 | 26 | 596 | 28 | 521 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1WKY_A | 6.17e-138 | 39 | 333 | 9 | 304 | Crystalstructure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module [Bacillus sp. JAMB-602] |
| 2WHJ_A | 6.54e-134 | 39 | 352 | 2 | 308 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 3JUG_A | 1.07e-131 | 22 | 333 | 11 | 319 | Crystalstructure of endo-beta-1,4-mannanase from the alkaliphilic Bacillus sp. N16-5 [Bacillus sp. N16-5] |
| 2WHL_A | 1.22e-127 | 39 | 332 | 1 | 293 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 1BQC_A | 1.67e-75 | 39 | 332 | 3 | 301 | Beta-MannanaseFrom Thermomonospora Fusca [Thermobifida fusca],2MAN_A Mannotriose Complex Of Thermomonospora Fusca Beta-Mannanase [Thermobifida fusca],3MAN_A Mannohexaose Complex Of Thermomonospora Fusca Beta-mannanase [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G1K3N4 | 3.58e-133 | 39 | 352 | 2 | 308 | Mannan endo-1,4-beta-mannosidase OS=Salipaludibacillus agaradhaerens OX=76935 PE=1 SV=1 |
| P22533 | 3.67e-81 | 59 | 596 | 55 | 515 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
| P51529 | 8.45e-77 | 27 | 354 | 27 | 347 | Mannan endo-1,4-beta-mannosidase OS=Streptomyces lividans OX=1916 GN=manA PE=1 SV=2 |
| B3PF24 | 1.41e-73 | 32 | 332 | 41 | 345 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=man5A PE=1 SV=1 |
| P29718 | 6.86e-70 | 452 | 596 | 1 | 145 | Uncharacterized protein in celA 5'region (Fragment) OS=Paenibacillus lautus OX=1401 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
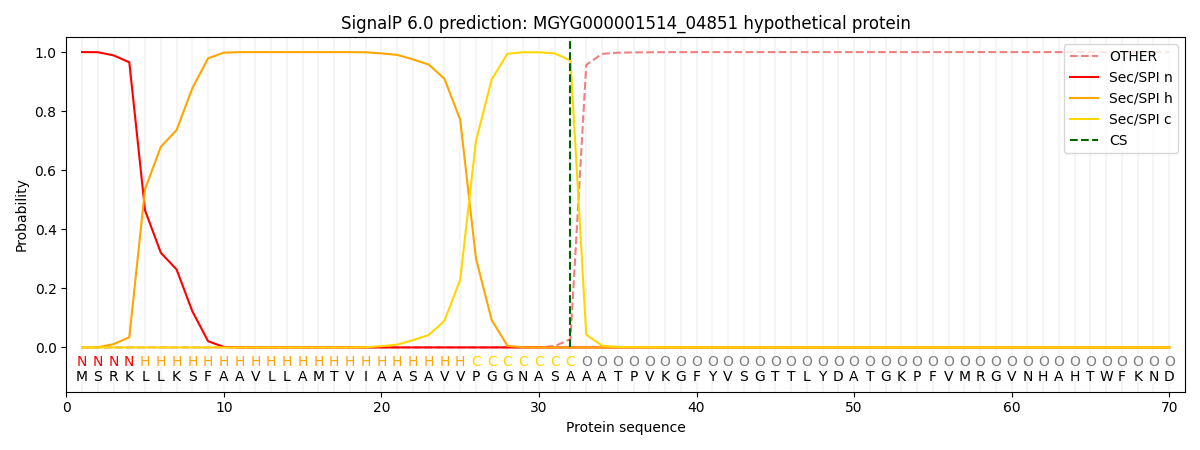
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.998978 | 0.000178 | 0.000208 | 0.000176 | 0.000149 |

