You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001508_01302
You are here: Home > Sequence: MGYG000001508_01302
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Thalassobacillus devorans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Halobacillaceae; Thalassobacillus; Thalassobacillus devorans | |||||||||||
| CAZyme ID | MGYG000001508_01302 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 405696; End: 409820 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 890 | 1261 | 3.4e-120 | 0.9786666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07523 | Big_3 | 2.08e-09 | 407 | 472 | 2 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam07523 | Big_3 | 5.12e-09 | 235 | 301 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam07523 | Big_3 | 1.28e-06 | 484 | 555 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam13229 | Beta_helix | 0.001 | 1001 | 1180 | 7 | 149 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam07523 | Big_3 | 0.004 | 314 | 393 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAS52182.1 | 0.0 | 11 | 1374 | 11 | 1375 |
| QTM99149.1 | 0.0 | 1 | 1374 | 1 | 1374 |
| BAB04213.1 | 0.0 | 6 | 1374 | 6 | 1376 |
| ADD02972.1 | 0.0 | 21 | 1301 | 22 | 1309 |
| ARK30471.1 | 0.0 | 18 | 1373 | 219 | 1602 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 3.76e-33 | 894 | 1263 | 18 | 374 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 4FDW_A | 2.77e-06 | 379 | 487 | 11 | 113 | Crystalstructure of a putative cell surface protein (BACOVA_01565) from Bacteroides ovatus ATCC 8483 at 2.05 A resolution [Bacteroides ovatus ATCC 8483] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 1.52e-67 | 561 | 1214 | 32 | 687 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A7 | 3.24e-32 | 894 | 1263 | 43 | 399 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P0C1A6 | 5.88e-32 | 894 | 1250 | 43 | 386 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| A0A401ETL2 | 4.70e-07 | 31 | 473 | 1004 | 1452 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
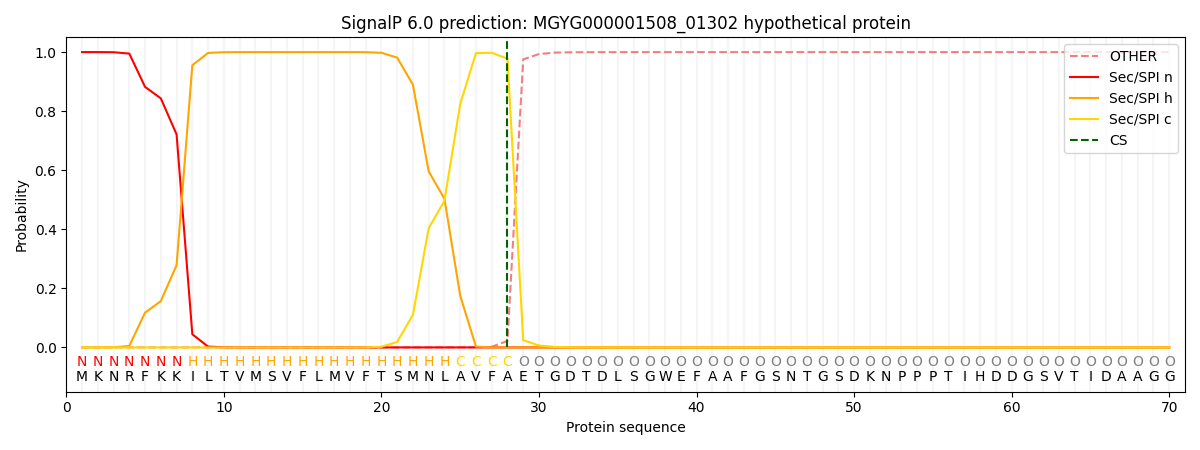
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000214 | 0.999096 | 0.000200 | 0.000178 | 0.000162 | 0.000152 |

