You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001470_00771
You are here: Home > Sequence: MGYG000001470_00771
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Virgibacillus kapii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Virgibacillus; Virgibacillus kapii | |||||||||||
| CAZyme ID | MGYG000001470_00771 | |||||||||||
| CAZy Family | AA10 | |||||||||||
| CAZyme Description | GlcNAc-binding protein A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 815592; End: 816449 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA10 | 34 | 195 | 2.2e-51 | 0.9943820224719101 |
| CBM5 | 219 | 258 | 3.1e-19 | 0.975 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13211 | PRK13211 | 5.81e-91 | 9 | 198 | 4 | 195 | N-acetylglucosamine-binding protein GbpA. |
| COG3397 | COG3397 | 5.19e-77 | 9 | 274 | 3 | 280 | Predicted carbohydrate-binding protein, contains CBM5 and CBM33 domains [General function prediction only]. |
| cd21177 | LPMO_AA10 | 1.13e-66 | 34 | 195 | 1 | 180 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
| pfam03067 | LPMO_10 | 3.74e-54 | 34 | 193 | 1 | 185 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| cd12215 | ChiC_BD | 2.94e-17 | 220 | 260 | 1 | 42 | Chitin-binding domain of chitinase C. Chitin-binding domain of chitinase C (ChiC) of Streptomyces griseus and related proteins. Chitinase C is a family 19 chitinase, and consists of a N-terminal chitin binding domain and a C-terminal chitin-catalytic domain that effects degradation. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. ChiC contains the characteristic chitin-binding aromatic residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRZ18344.1 | 3.62e-183 | 1 | 285 | 1 | 281 |
| AUJ23668.1 | 7.08e-141 | 11 | 276 | 6 | 269 |
| SQI51376.1 | 2.49e-121 | 9 | 262 | 9 | 269 |
| ARD48446.1 | 1.95e-96 | 13 | 261 | 13 | 279 |
| ARK21810.1 | 2.93e-96 | 11 | 265 | 11 | 285 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WSZ_A | 2.37e-61 | 34 | 195 | 1 | 165 | Crystalstructure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_B Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_C Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_D Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki] |
| 5LW4_A | 2.26e-54 | 34 | 195 | 1 | 168 | NMRsolution structure of the apo-form of the chitin-active lytic polysaccharide monooxygenase BlLPMO10A [Bacillus licheniformis],6TWE_A Cu(I) NMR solution structure of the chitin-active lytic polysaccharide monooxygenase BlLPMO10A [Bacillus licheniformis DSM 13 = ATCC 14580] |
| 2XWX_A | 4.20e-54 | 34 | 273 | 1 | 239 | Vibriocholerae colonization factor GbpA crystal structure [Vibrio cholerae],2XWX_B Vibrio cholerae colonization factor GbpA crystal structure [Vibrio cholerae] |
| 5L2V_A | 7.48e-54 | 34 | 195 | 1 | 163 | Catalyticdomain of LPMO Lmo2467 from Listeria monocytogenes [Listeria monocytogenes 10403S],5L2V_B Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes [Listeria monocytogenes 10403S] |
| 2BEM_A | 1.92e-52 | 34 | 195 | 1 | 167 | Crystalstructure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2BEM_B Crystal structure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2BEM_C Crystal structure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2LHS_A Structure of the chitin binding protein 21 (CBP21) [Serratia marcescens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8EHY2 | 1.36e-58 | 8 | 205 | 2 | 204 | GlcNAc-binding protein A OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=gbpA PE=3 SV=2 |
| Q6LKG5 | 1.47e-56 | 27 | 199 | 18 | 205 | GlcNAc-binding protein A OS=Photobacterium profundum (strain SS9) OX=298386 GN=gbpA PE=3 SV=1 |
| Q5E183 | 1.70e-55 | 22 | 198 | 12 | 206 | GlcNAc-binding protein A OS=Aliivibrio fischeri (strain ATCC 700601 / ES114) OX=312309 GN=gbpA PE=3 SV=1 |
| B5ESR7 | 1.70e-55 | 22 | 198 | 12 | 206 | GlcNAc-binding protein A OS=Aliivibrio fischeri (strain MJ11) OX=388396 GN=gbpA PE=3 SV=1 |
| Q87FT0 | 1.17e-54 | 22 | 199 | 12 | 204 | GlcNAc-binding protein A OS=Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633) OX=223926 GN=gbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
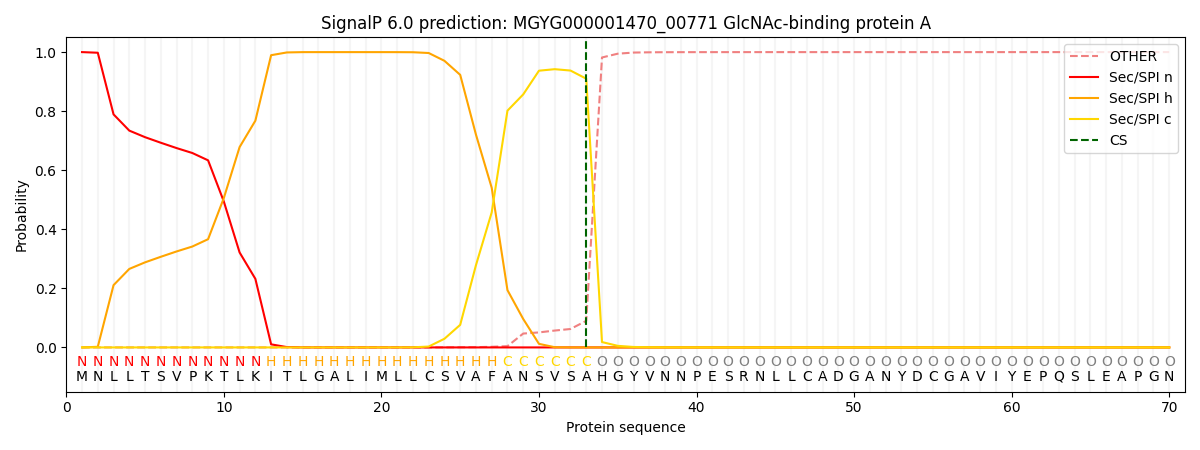
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000217 | 0.999193 | 0.000155 | 0.000155 | 0.000136 | 0.000125 |

