You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001468_01867
You are here: Home > Sequence: MGYG000001468_01867
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Blastococcus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Geodermatophilaceae; Blastococcus; Blastococcus massiliensis | |||||||||||
| CAZyme ID | MGYG000001468_01867 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 163172; End: 165922 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01804 | Penicil_amidase | 5.43e-83 | 124 | 812 | 1 | 524 | Penicillin amidase. Penicillin amidase or penicillin acylase EC:3.5.1.11 catalyzes the hydrolysis of benzylpenicillin to phenylacetic acid and 6-aminopenicillanic acid (6-APA) a key intermediate in the the synthesis of penicillins. Also in the family is cephalosporin acylase and aculeacin A acylase which are involved in the synthesis of related peptide antibiotics. |
| COG2366 | PvdQ | 1.87e-44 | 124 | 896 | 42 | 679 | Acyl-homoserine lactone (AHL) acylase PvdQ [Secondary metabolites biosynthesis, transport and catabolism]. |
| cd03748 | Ntn_PGA | 2.10e-41 | 397 | 775 | 1 | 356 | Penicillin G acylase (PGA) is the key enzyme in the industrial production of beta-lactam antibiotics. PGA hydrolyzes the side chain of penicillin G and related beta-lactam antibiotics releasing 6-amino penicillanic acid (6-APA), a building block in the production of semisynthetic penicillins. PGA is widely distributed among microorganisms, including bacteria, yeast and filamentous fungi but it's in vivo role remains unclear. |
| cd03747 | Ntn_PGA_like | 6.29e-40 | 397 | 706 | 1 | 291 | Penicillin G acylase (PGA) belongs to a family of beta-lactam acylases that includes cephalosporin acylase (CA) and aculeacin A acylase. PGA and CA are crucial for the production of backbone chemicals like 6-aminopenicillanic acid and 7-aminocephalosporanic acid (7-ACA), which can be used to synthesize semi-synthetic penicillins and cephalosporins, respectively. While both PGA and CA have a conserved Ntn (N-terminal nucleophile) hydrolase fold and the structural similarity at their active sites is very high, their sequence similarity is low. |
| cd01936 | Ntn_CA | 1.62e-23 | 389 | 771 | 50 | 421 | Cephalosporin acylase (CA) belongs to a family of beta-lactam acylases that includes penicillin G acylase (PGA) and aculeacin A acylase. PGA and CA are crucial for the production of backbone chemicals like 6-aminopenicillanic acid and 7-aminocephalosporanic acid (7-ACA), which can be used to synthesize semi-synthetic penicillins and cephalosporins, respectively. While both PGA and CA have a conserved Ntn (N-terminal nucleophile) hydrolase fold and the structural similarity at their active sites is very high, their sequence similarity to other Ntn's is low. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADB30173.1 | 4.05e-173 | 5 | 914 | 20 | 938 |
| QKW18046.1 | 2.94e-167 | 33 | 914 | 50 | 933 |
| QNE20646.1 | 6.74e-165 | 5 | 914 | 46 | 961 |
| BAJ26473.1 | 7.75e-162 | 33 | 914 | 50 | 930 |
| QFZ16597.1 | 2.72e-161 | 33 | 914 | 43 | 927 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6NVY_B | 2.55e-35 | 397 | 804 | 1 | 380 | Crystalstructure of penicillin G acylase from Bacillus thermotolerans [Quasibacillus thermotolerans],6NVY_D Crystal structure of penicillin G acylase from Bacillus thermotolerans [Quasibacillus thermotolerans] |
| 6NVX_B | 3.06e-29 | 397 | 804 | 1 | 380 | Crystalstructure of penicillin G acylase from Bacillus sp. FJAT-27231 [Bacillus sp. FJAT-27231] |
| 7REP_A | 1.68e-17 | 397 | 781 | 1 | 363 | ChainA, Penicillin G acylase [Kluyvera cryocrescens] |
| 1CP9_B | 7.54e-17 | 397 | 794 | 1 | 363 | CrystalStructure Of Penicillin G Acylase From The Bro1 Mutant Strain Of Providencia Rettgeri [Providencia rettgeri] |
| 4PEL_B | 1.37e-16 | 398 | 781 | 2 | 363 | S1Cmutant of Penicillin G acylase from Kluyvera citrophila [Kluyvera cryocrescens],4PEL_D S1C mutant of Penicillin G acylase from Kluyvera citrophila [Kluyvera cryocrescens],4PEL_F S1C mutant of Penicillin G acylase from Kluyvera citrophila [Kluyvera cryocrescens],4PEL_H S1C mutant of Penicillin G acylase from Kluyvera citrophila [Kluyvera cryocrescens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P31956 | 1.71e-44 | 118 | 817 | 26 | 657 | Penicillin G acylase OS=Rhizobium viscosum OX=1673 GN=pac PE=1 SV=1 |
| Q60136 | 1.30e-42 | 118 | 804 | 26 | 645 | Penicillin G acylase OS=Priestia megaterium OX=1404 GN=pac PE=1 SV=1 |
| P07941 | 5.96e-29 | 120 | 781 | 29 | 650 | Penicillin G acylase OS=Kluyvera cryocrescens OX=580 GN=pac PE=1 SV=1 |
| P06875 | 2.45e-25 | 118 | 685 | 27 | 559 | Penicillin G acylase OS=Escherichia coli OX=562 GN=pac PE=1 SV=2 |
| Q3KH00 | 1.98e-15 | 122 | 781 | 51 | 614 | Acyl-homoserine lactone acylase QuiP OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=quiP PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
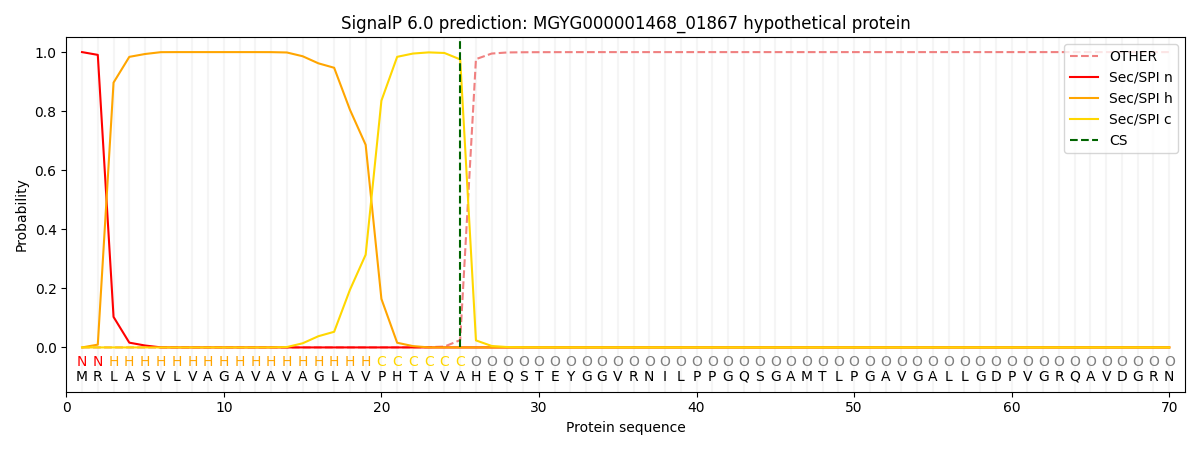
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000230 | 0.999121 | 0.000160 | 0.000171 | 0.000153 | 0.000139 |
