You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_01926
You are here: Home > Sequence: MGYG000001433_01926
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_01926 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2517710; End: 2520847 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 611 | 927 | 4.9e-67 | 0.9929078014184397 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.82e-29 | 643 | 922 | 33 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 8.59e-14 | 638 | 921 | 77 | 360 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam18962 | Por_Secre_tail | 4.30e-13 | 974 | 1043 | 1 | 72 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
| TIGR04183 | Por_Secre_tail | 3.24e-10 | 974 | 1044 | 1 | 71 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| NF033708 | T9SS_Cterm_ChiA | 8.59e-06 | 991 | 1043 | 1 | 54 | T9SS sorting signal type C. The sorting signals of type IX secretion systems (T9SS) in the CFB bacteria are long, compared to other prokaryotic C-terminal sorting motif-containing signals, including LPXTG, PEP-CTERM, and GlyGly-TERM, and they seem to contain multiple motifs. A few T9SS substrates, including ChiA, have a variant form of T9SS sorting signal that may score poorly to both TIGR04183 (type A) and TIGR04131 (type B), depend on T9SS for secretion, but are released from the cell rather than left anchored to the cell surface. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI63368.1 | 8.95e-151 | 30 | 1045 | 530 | 1467 |
| QQY82914.1 | 1.97e-80 | 599 | 1044 | 13 | 420 |
| AUS05313.1 | 3.41e-78 | 609 | 1028 | 32 | 418 |
| QUT26200.1 | 2.00e-75 | 605 | 950 | 33 | 356 |
| ALJ47659.1 | 2.00e-75 | 605 | 950 | 33 | 356 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4LX4_A | 2.18e-46 | 610 | 958 | 12 | 321 | CrystalStructure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_B Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_C Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_D Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],6R2J_A Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_B Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_C Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_D Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501] |
| 4EE9_A | 1.08e-43 | 612 | 933 | 5 | 291 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
| 7P6G_A | 2.70e-43 | 612 | 933 | 5 | 291 | ChainA, Endoglucanase [uncultured bacterium],7P6G_B Chain B, Endoglucanase [uncultured bacterium],7P6H_A Chain A, Endoglucanase [uncultured bacterium],7P6H_B Chain B, Endoglucanase [uncultured bacterium] |
| 6ZZ3_A | 3.57e-43 | 612 | 933 | 5 | 291 | ChainA, Endoglucanase [uncultured bacterium],6ZZ3_B Chain B, Endoglucanase [uncultured bacterium],6ZZ3_C Chain C, Endoglucanase [uncultured bacterium],6ZZ3_D Chain D, Endoglucanase [uncultured bacterium] |
| 7P6I_A | 3.67e-43 | 612 | 933 | 5 | 291 | ChainA, Endoglucanase [uncultured bacterium],7P6J_A Chain A, Endoglucanase [uncultured bacterium],7P6J_B Chain B, Endoglucanase [uncultured bacterium],7P6J_C Chain C, Endoglucanase [uncultured bacterium],7P6J_D Chain D, Endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q1HFS8 | 4.44e-33 | 611 | 951 | 29 | 327 | Endo-beta-1,4-glucanase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglB PE=1 SV=1 |
| P58599 | 7.81e-31 | 611 | 952 | 117 | 424 | Endoglucanase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=egl PE=3 SV=1 |
| P17974 | 8.07e-31 | 611 | 952 | 119 | 426 | Endoglucanase OS=Ralstonia solanacearum OX=305 GN=egl PE=1 SV=2 |
| O74706 | 2.39e-26 | 611 | 951 | 34 | 331 | Endo-beta-1,4-glucanase B OS=Aspergillus niger OX=5061 GN=eglB PE=1 SV=1 |
| A2QPC3 | 2.39e-26 | 611 | 951 | 34 | 331 | Probable endo-beta-1,4-glucanase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
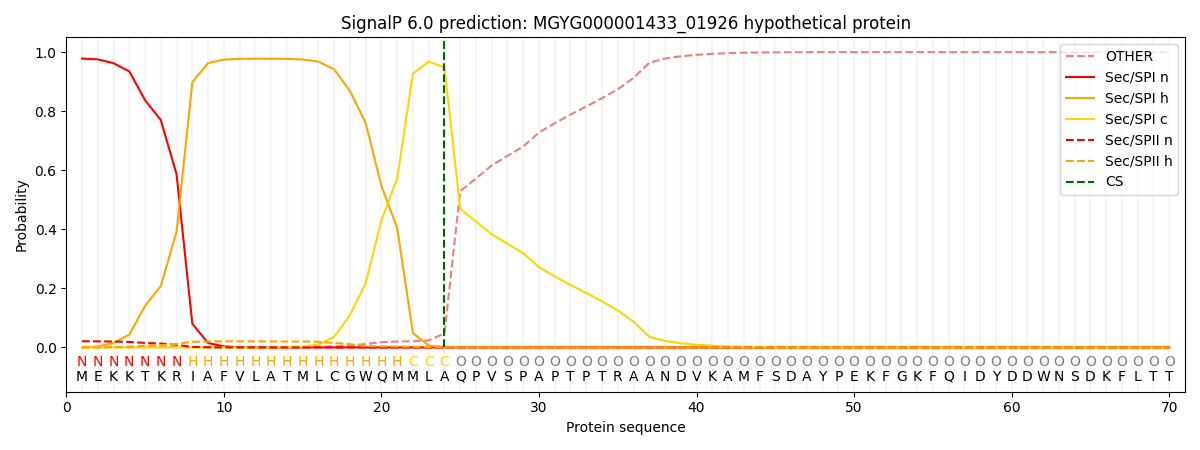
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002607 | 0.974130 | 0.022483 | 0.000293 | 0.000242 | 0.000224 |
