You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_01483
You are here: Home > Sequence: MGYG000001422_01483
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_01483 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1687199; End: 1689352 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 452 | 711 | 4.4e-44 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 5.16e-28 | 452 | 710 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.33e-27 | 452 | 712 | 96 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 6.20e-17 | 459 | 717 | 126 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.19e-09 | 55 | 138 | 1 | 85 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.22e-04 | 97 | 139 | 1 | 43 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ61518.1 | 0.0 | 1 | 717 | 1 | 721 |
| AXH21505.1 | 0.0 | 1 | 717 | 1 | 746 |
| EDV05072.1 | 0.0 | 1 | 717 | 1 | 746 |
| QUT92912.1 | 7.34e-264 | 1 | 717 | 1 | 730 |
| QDO69412.1 | 5.08e-226 | 1 | 717 | 8 | 742 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGS_A | 7.16e-63 | 156 | 300 | 2 | 150 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4QPW_A | 3.04e-59 | 161 | 298 | 1 | 142 | BiXyn10ACBM1 with Xylohexaose Bound [Bacteroides intestinalis DSM 17393] |
| 1I1W_A | 7.03e-14 | 453 | 714 | 101 | 301 | 0.89AUltra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
| 2BNJ_A | 1.27e-13 | 453 | 714 | 101 | 301 | Thexylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. [Thermoascus aurantiacus] |
| 1TUX_A | 5.41e-13 | 453 | 707 | 101 | 294 | HighResolution Crystal Structure Of A Thermostable Xylanase From Thermoascus Aurantiacus [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O59859 | 3.21e-16 | 453 | 714 | 127 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
| W0HFK8 | 6.42e-15 | 453 | 714 | 131 | 331 | Endo-1,4-beta-xylanase 1 OS=Rhizopus oryzae OX=64495 GN=xyn1 PE=1 SV=1 |
| Q6PRW6 | 6.26e-14 | 453 | 707 | 131 | 324 | Endo-1,4-beta-xylanase OS=Penicillium chrysogenum OX=5076 GN=Xyn PE=1 SV=1 |
| P29417 | 2.00e-13 | 453 | 707 | 131 | 324 | Endo-1,4-beta-xylanase OS=Penicillium chrysogenum OX=5076 GN=XYLP PE=1 SV=2 |
| P23360 | 7.08e-12 | 453 | 714 | 127 | 327 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
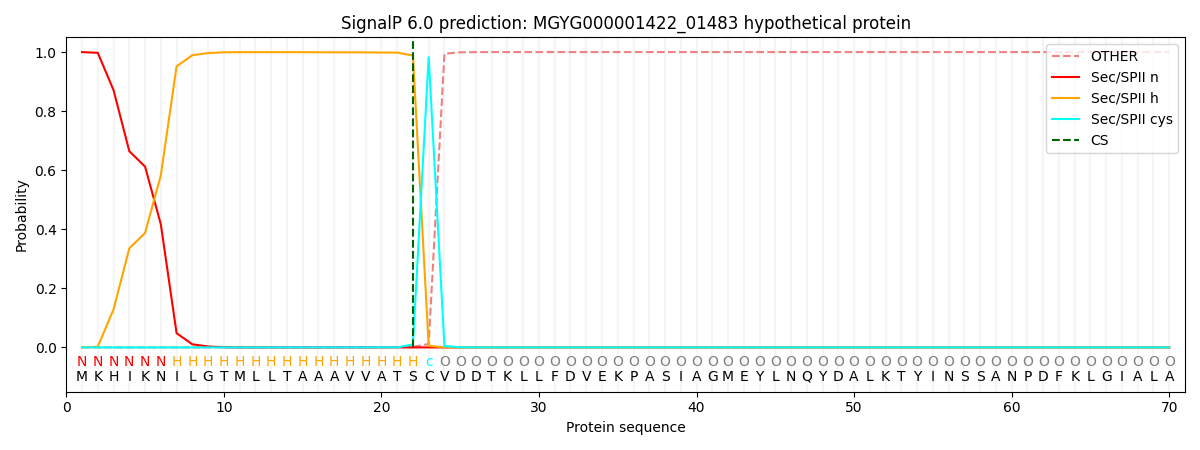
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000005 | 0.000083 | 0.999941 | 0.000000 | 0.000000 | 0.000000 |
