You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_01480
You are here: Home > Sequence: MGYG000001422_01480
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_01480 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1681307; End: 1683280 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 69 | 352 | 1.6e-111 | 0.9884169884169884 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.03e-09 | 109 | 309 | 34 | 208 | Cellulase (glycosyl hydrolase family 5). |
| pfam02368 | Big_2 | 1.23e-04 | 452 | 524 | 12 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| COG2730 | BglC | 2.24e-04 | 77 | 275 | 37 | 234 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| smart00635 | BID_2 | 0.001 | 460 | 518 | 20 | 79 | Bacterial Ig-like domain 2. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ61520.1 | 0.0 | 1 | 657 | 1 | 655 |
| QUT92910.1 | 0.0 | 1 | 657 | 1 | 653 |
| EEF88964.1 | 0.0 | 2 | 657 | 13 | 666 |
| QDO69414.1 | 0.0 | 6 | 657 | 2 | 656 |
| EDV05070.1 | 0.0 | 1 | 657 | 1 | 660 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
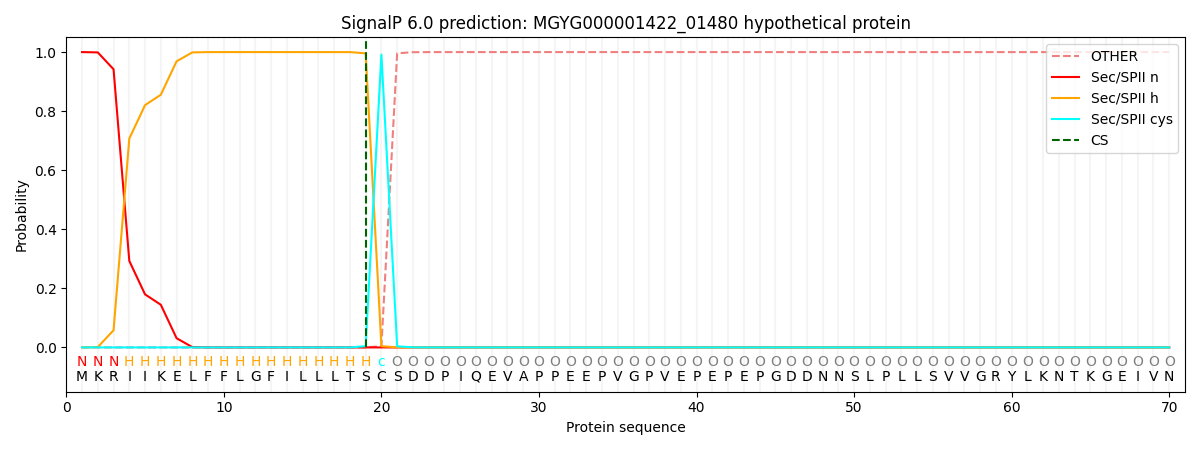
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000057 | 0.000000 | 0.000000 | 0.000000 |
