You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001390_01893
You are here: Home > Sequence: MGYG000001390_01893
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus_O smithii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; Domibacillaceae; Bacillus_O; Bacillus_O smithii | |||||||||||
| CAZyme ID | MGYG000001390_01893 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | putative peptidoglycan endopeptidase LytE | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 82966; End: 83991 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM50 | 94 | 135 | 4.3e-18 | 0.95 |
| CBM50 | 164 | 206 | 4.9e-17 | 0.975 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 1.63e-38 | 235 | 339 | 1 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK13914 | PRK13914 | 1.53e-37 | 96 | 339 | 204 | 480 | invasion associated endopeptidase. |
| COG0791 | Spr | 5.87e-37 | 206 | 338 | 57 | 197 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| PRK06347 | PRK06347 | 1.81e-31 | 15 | 225 | 321 | 535 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 3.03e-30 | 29 | 205 | 408 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKP45810.1 | 7.32e-212 | 1 | 341 | 1 | 341 |
| QOK25940.1 | 1.81e-98 | 1 | 341 | 1 | 345 |
| BCB05878.1 | 1.29e-97 | 1 | 341 | 1 | 350 |
| AND42279.1 | 2.58e-97 | 1 | 341 | 1 | 341 |
| SQI62971.1 | 2.03e-95 | 1 | 341 | 1 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HPE_A | 1.20e-22 | 214 | 333 | 178 | 299 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 4FDY_A | 1.11e-20 | 229 | 333 | 197 | 303 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
| 4XCM_A | 2.45e-20 | 92 | 331 | 3 | 221 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
| 7CFL_A | 1.09e-18 | 225 | 339 | 16 | 136 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 6B8C_A | 2.56e-16 | 219 | 325 | 24 | 131 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54421 | 1.66e-75 | 1 | 340 | 1 | 334 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| O31852 | 3.50e-63 | 29 | 334 | 90 | 407 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
| O07532 | 5.32e-63 | 29 | 341 | 176 | 488 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
| Q01837 | 9.63e-36 | 92 | 328 | 196 | 512 | Probable endopeptidase p60 OS=Listeria ivanovii OX=1638 GN=iap PE=3 SV=1 |
| Q01839 | 5.80e-33 | 93 | 328 | 197 | 512 | Probable endopeptidase p60 OS=Listeria welshimeri OX=1643 GN=iap PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
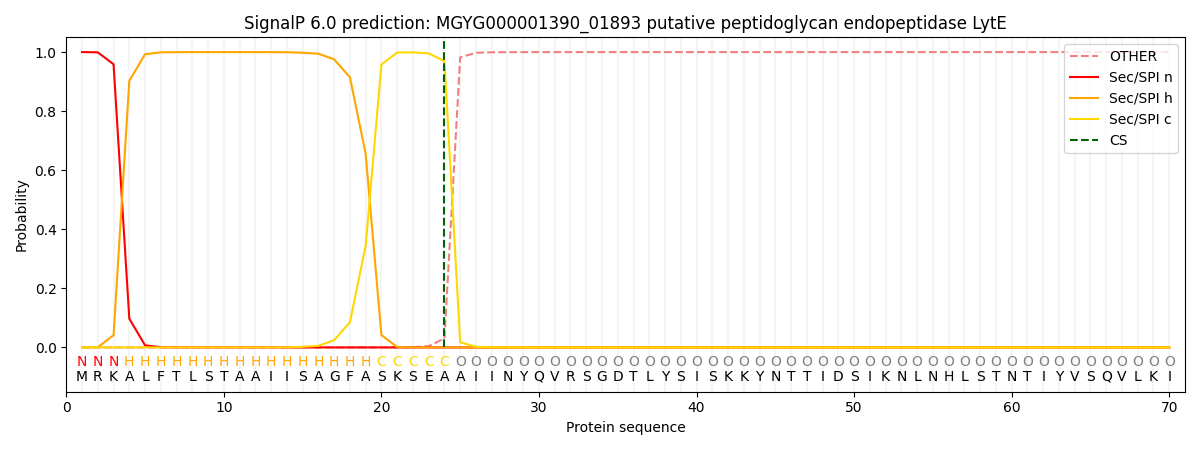
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000221 | 0.999058 | 0.000212 | 0.000182 | 0.000164 | 0.000140 |
