You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001378_04497
You are here: Home > Sequence: MGYG000001378_04497
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
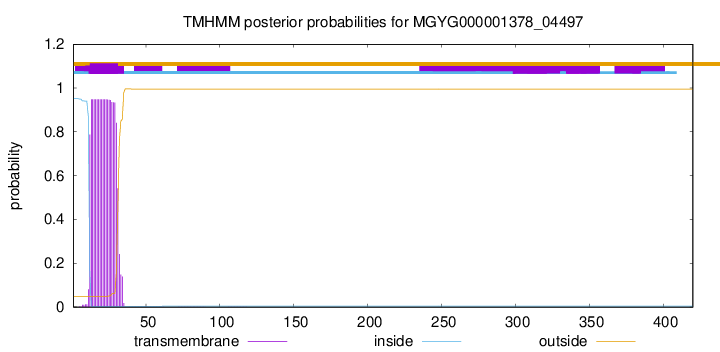
TMHMM annotations
Basic Information help
| Species | Bacteroides ovatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides ovatus | |||||||||||
| CAZyme ID | MGYG000001378_04497 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 137308; End: 138570 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 294 | 406 | 2.5e-19 | 0.8225806451612904 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 8.09e-18 | 292 | 414 | 6 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 3.02e-11 | 304 | 416 | 29 | 143 | Substituted updates: Jan 31, 2002 |
| pfam18911 | PKD_4 | 4.21e-11 | 182 | 254 | 3 | 81 | PKD domain. This entry is composed of PKD domains found in bacterial surface proteins. |
| smart00231 | FA58C | 1.72e-09 | 300 | 417 | 24 | 139 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
| smart00089 | PKD | 2.43e-09 | 196 | 260 | 2 | 73 | Repeats in polycystic kidney disease 1 (PKD1) and other proteins. Polycystic kidney disease 1 protein contains 14 repeats, present elsewhere such as in microbial collagenases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT72072.1 | 4.92e-313 | 1 | 420 | 16 | 435 |
| QNL39441.1 | 1.20e-298 | 1 | 420 | 1 | 420 |
| BCI63788.1 | 2.74e-135 | 1 | 417 | 1 | 418 |
| QHS63755.1 | 2.40e-21 | 257 | 415 | 165 | 322 |
| AWW31575.1 | 7.16e-21 | 244 | 416 | 194 | 355 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3F2Z_A | 1.70e-14 | 270 | 419 | 4 | 150 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
| 1WCQ_A | 2.30e-14 | 242 | 416 | 432 | 599 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BZD_A | 2.30e-14 | 242 | 416 | 432 | 599 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
| 2BER_A | 2.30e-14 | 242 | 416 | 432 | 599 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
| 1W8N_A | 2.30e-14 | 242 | 416 | 432 | 599 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 1.33e-13 | 242 | 416 | 478 | 645 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| Q0TR53 | 3.73e-07 | 300 | 416 | 645 | 767 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 3.73e-07 | 300 | 416 | 645 | 767 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q46085 | 1.99e-06 | 188 | 271 | 722 | 816 | Collagenase ColH OS=Hathewaya histolytica OX=1498 GN=colH PE=1 SV=1 |
| Q58863 | 2.62e-06 | 106 | 266 | 799 | 973 | Uncharacterized protein MJ1468 OS=Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) OX=243232 GN=MJ1468 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
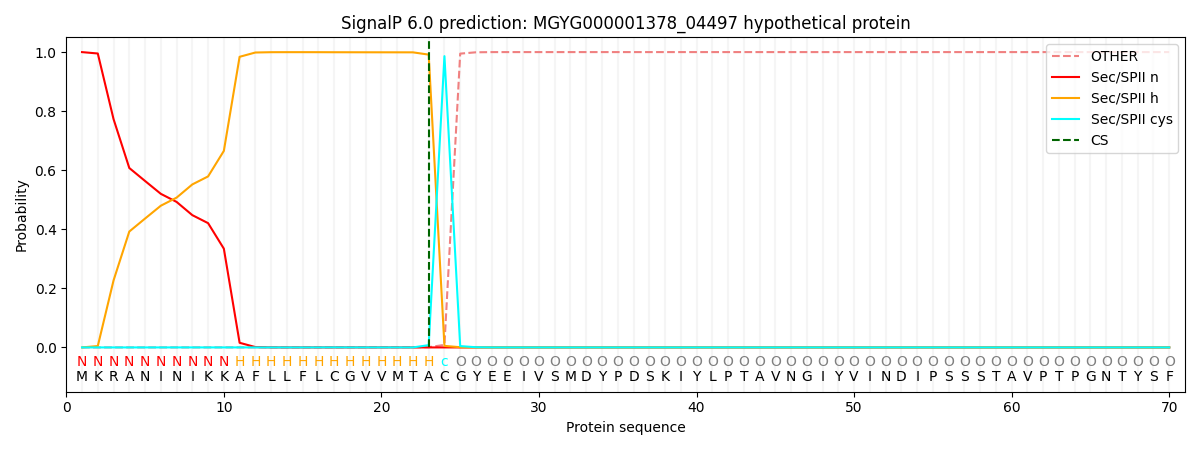
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000015 | 1.000015 | 0.000000 | 0.000000 | 0.000000 |