You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001374_02062
You are here: Home > Sequence: MGYG000001374_02062
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter torques | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter torques | |||||||||||
| CAZyme ID | MGYG000001374_02062 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 271760; End: 276664 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 191 | 484 | 4.7e-71 | 0.9898305084745763 |
| CBM32 | 1224 | 1329 | 1.2e-17 | 0.7419354838709677 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 2.18e-112 | 191 | 482 | 1 | 291 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 1.58e-16 | 43 | 184 | 1 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 2.51e-15 | 1213 | 1332 | 1 | 117 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| NF033201 | Vip_LPXTG_Lm | 1.09e-09 | 1421 | 1585 | 250 | 394 | cell invasion LPXTG protein Vip. Vip (Virulence protein), like the LPXTG-type internalins, is an LPXTG-anchored surface protein of the mammalian cell-invading pathogen Listeria monocytogenes, but absent from the related species Listeria innocua. For certain cell types, Vip is required for Listeria's ability to invade. It appears to bind the endoplasmic reticulum (ER) resident chaperone Gp96 as its receptor. |
| PTZ00449 | PTZ00449 | 6.83e-09 | 1451 | 1510 | 588 | 653 | 104 kDa microneme/rhoptry antigen; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASM68920.1 | 4.61e-278 | 35 | 1450 | 48 | 1325 |
| AIY84741.1 | 8.19e-199 | 10 | 1165 | 2 | 1164 |
| APC49529.1 | 8.16e-197 | 16 | 1165 | 7 | 1145 |
| APC49528.1 | 1.16e-190 | 71 | 1165 | 3 | 1088 |
| ASK62202.1 | 5.82e-190 | 16 | 1185 | 7 | 1175 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PWI_A | 1.48e-101 | 36 | 630 | 26 | 617 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 6PV4_A | 8.31e-85 | 34 | 630 | 22 | 637 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 6PV5_A | 1.80e-66 | 26 | 632 | 21 | 618 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 5MI4_A | 3.47e-55 | 97 | 636 | 53 | 582 | BtGH84mutant with covalent modification by MA3 [Bacteroides thetaiotaomicron VPI-5482],5MI5_A BtGH84 mutant with covalent modification by MA3 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482],5MI6_A BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G [Bacteroides thetaiotaomicron VPI-5482],5MI7_A BtGH84 mutant with covalent modification by MA4 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482] |
| 2J4G_A | 9.44e-55 | 97 | 636 | 41 | 570 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482],2J4G_B Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 3.03e-114 | 11 | 1167 | 7 | 1350 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q89ZI2 | 7.07e-54 | 97 | 636 | 63 | 592 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q0TR53 | 5.54e-45 | 37 | 873 | 34 | 826 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 5.54e-45 | 37 | 873 | 34 | 826 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
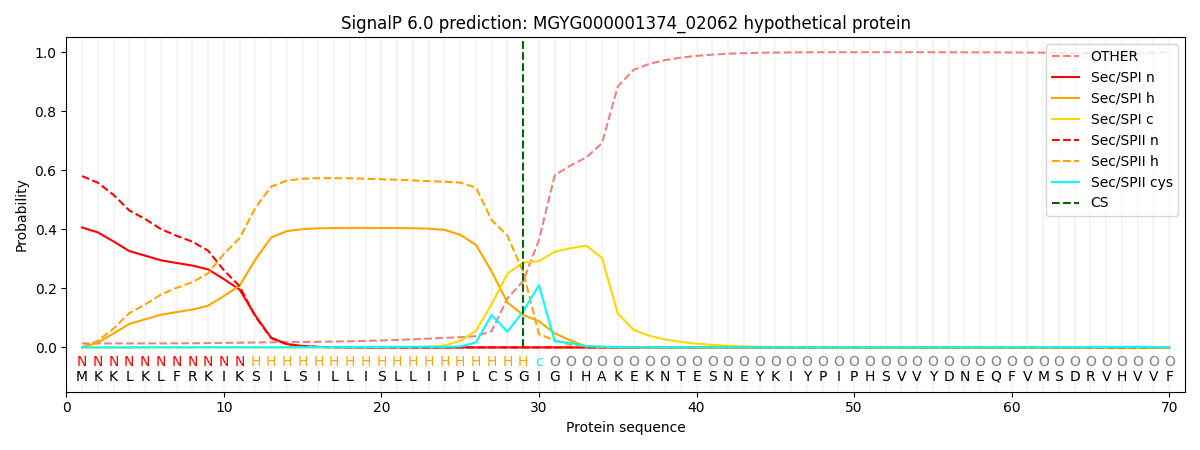
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.015761 | 0.395910 | 0.586924 | 0.000683 | 0.000367 | 0.000334 |

