You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001368_00164
You are here: Home > Sequence: MGYG000001368_00164
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
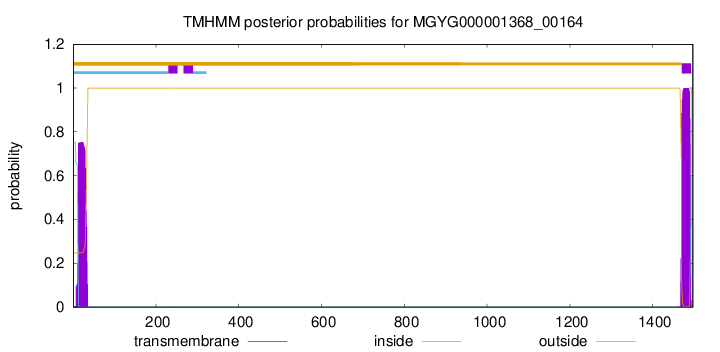
TMHMM annotations
Basic Information help
| Species | Muricomes fissicatena_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Muricomes; Muricomes fissicatena_A | |||||||||||
| CAZyme ID | MGYG000001368_00164 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 191830; End: 196323 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 438 | 1087 | 1.9e-52 | 0.8476454293628809 |
| CBM32 | 313 | 439 | 2.2e-17 | 0.8467741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 2.75e-15 | 303 | 439 | 4 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 3.20e-06 | 315 | 425 | 25 | 129 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 1.62e-04 | 173 | 274 | 25 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam07058 | MAP70 | 0.002 | 1205 | 1392 | 1 | 166 | Microtubule-associated protein 70. This family represents a family of plant microtubule-associated proteins of size 70 kDa. The proteins contain four predicted coiled-coil domains, and truncation studies identify a central domain that targets the proteins to microtubules. It has no predicted trans-membrane domains, and the region between the coils from approximately residues 240-483 is the targetting region. |
| smart00231 | FA58C | 0.003 | 313 | 425 | 22 | 124 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 5.69e-201 | 40 | 1135 | 42 | 1084 |
| QXE33659.1 | 6.27e-165 | 28 | 1132 | 51 | 952 |
| AWS40658.1 | 5.03e-158 | 42 | 1132 | 41 | 926 |
| BCB76148.1 | 4.05e-135 | 46 | 1132 | 1 | 879 |
| QKW17790.1 | 7.15e-86 | 443 | 1106 | 129 | 754 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 1.45e-11 | 558 | 1064 | 302 | 792 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
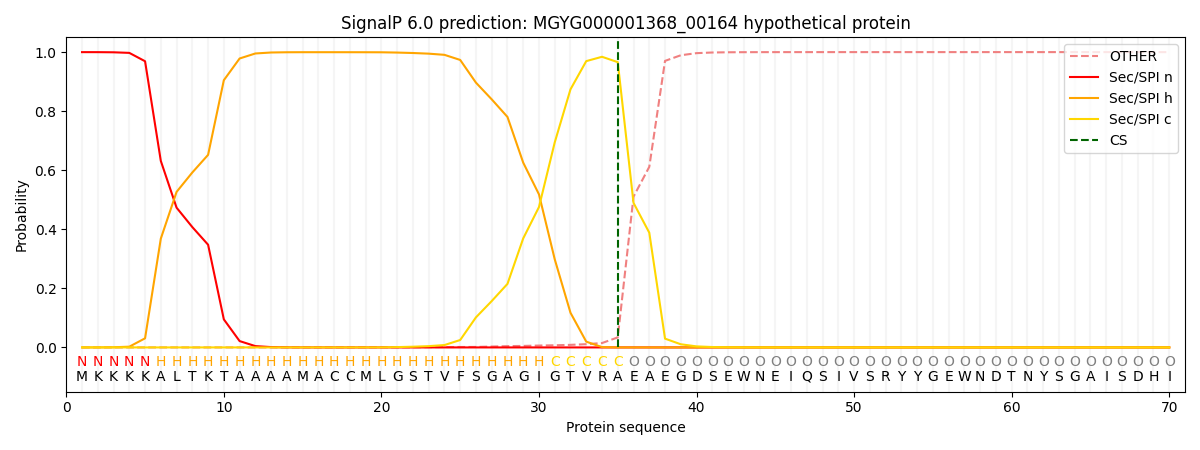
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000359 | 0.998622 | 0.000451 | 0.000216 | 0.000176 | 0.000146 |