You are browsing environment: HUMAN GUT
MGYG000001350_00804
Basic Information
help
Species
Fusobacterium periodonticum_B
Lineage
Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium; Fusobacterium periodonticum_B
CAZyme ID
MGYG000001350_00804
CAZy Family
GT21
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001350
2454675
Isolate
not provided
not provided
Gene Location
Start: 513094;
End: 514290
Strand: +
No EC number prediction in MGYG000001350_00804.
Family
Start
End
Evalue
family coverage
GT21
44
270
1.1e-21
0.9699570815450643
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam13506
Glyco_transf_21
1.49e-37
99
274
2
174
Glycosyl transferase family 21. This is a family of ceramide beta-glucosyltransferases - EC:2.4.1.80.
more
COG1215
BcsA
7.60e-06
35
384
48
385
Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility].
more
pfam13641
Glyco_tranf_2_3
5.95e-04
43
270
4
228
Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis.
more
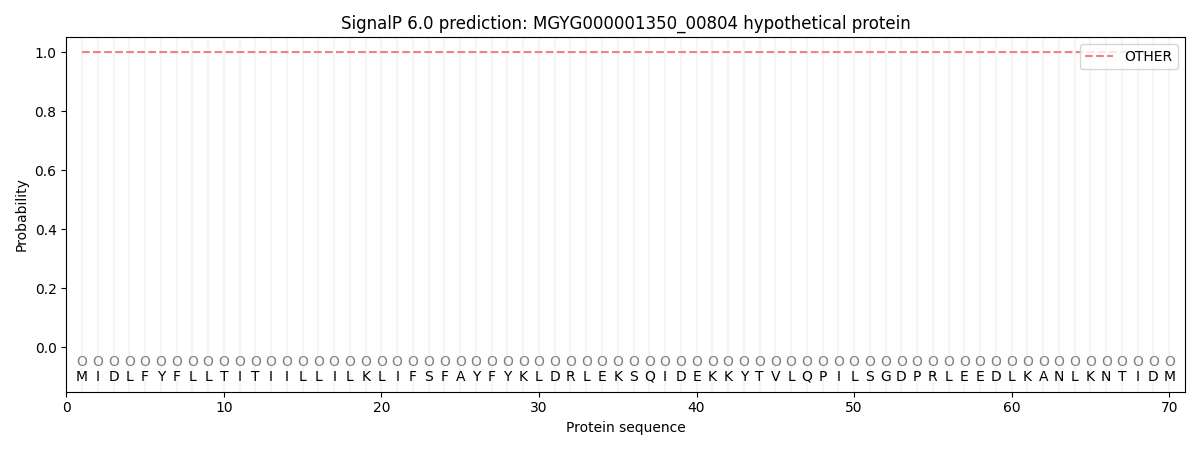
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000047
0.000001
0.000000
0.000000
0.000000
0.000000
start
end
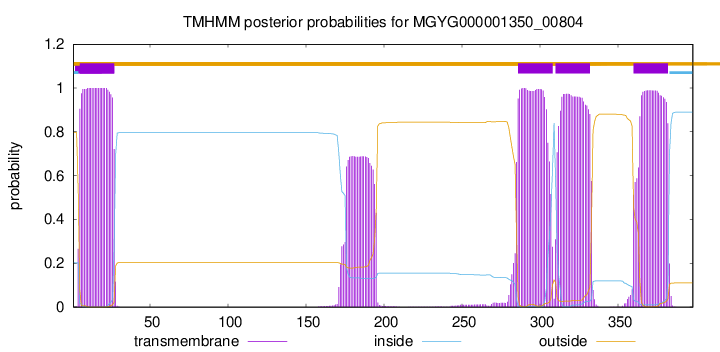
5
27
286
308
310
332
360
382