You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001328_01931
You are here: Home > Sequence: MGYG000001328_01931
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
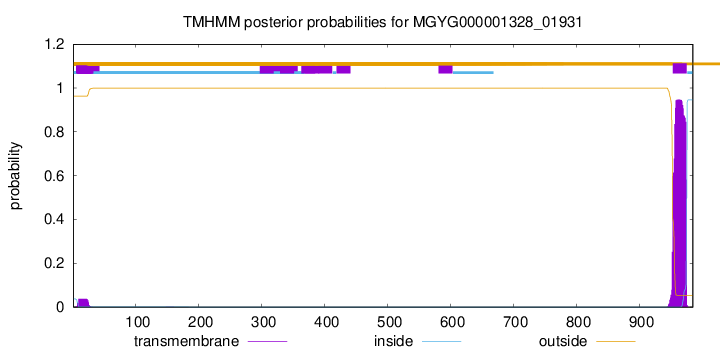
TMHMM annotations
Basic Information help
| Species | Clostridium tertium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium tertium | |||||||||||
| CAZyme ID | MGYG000001328_01931 | |||||||||||
| CAZy Family | GH171 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1679508; End: 1682465 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH171 | 58 | 421 | 4.7e-140 | 0.9971830985915493 |
| GH171 | 453 | 816 | 8.1e-137 | 0.9971830985915493 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07075 | DUF1343 | 0.0 | 454 | 816 | 1 | 362 | Protein of unknown function (DUF1343). This family consists of several hypothetical bacterial proteins of around 400 residues in length. The function of this family is unknown. |
| pfam07075 | DUF1343 | 0.0 | 59 | 421 | 1 | 362 | Protein of unknown function (DUF1343). This family consists of several hypothetical bacterial proteins of around 400 residues in length. The function of this family is unknown. |
| COG3876 | YbbC | 1.17e-167 | 16 | 421 | 8 | 409 | Uncharacterized conserved protein YbbC, DUF1343 family [Function unknown]. |
| COG3876 | YbbC | 2.02e-159 | 434 | 816 | 26 | 409 | Uncharacterized conserved protein YbbC, DUF1343 family [Function unknown]. |
| pfam16403 | DUF5011 | 6.72e-10 | 830 | 898 | 6 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVK18518.1 | 4.02e-143 | 39 | 422 | 2 | 380 |
| AJQ29948.1 | 1.28e-141 | 24 | 421 | 31 | 421 |
| AIF54236.1 | 4.90e-139 | 39 | 422 | 49 | 425 |
| SNX53310.1 | 4.09e-136 | 434 | 817 | 2 | 380 |
| ARP42412.1 | 1.96e-135 | 5 | 422 | 1 | 418 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4K05_A | 1.15e-75 | 434 | 816 | 13 | 398 | Crystalstructure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343],4K05_B Crystal structure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343] |
| 4JJA_A | 1.35e-49 | 39 | 424 | 6 | 371 | Crystalstructure of a DUF1343 family protein (BF0379) from Bacteroides fragilis NCTC 9343 at 1.30 A resolution [Bacteroides fragilis NCTC 9343] |
| 2KPN_A | 9.44e-11 | 834 | 899 | 20 | 84 | SolutionNMR structure of a Bacterial Ig-like (Big_3) domain from Bacillus cereus. Northeast Structural Genomics Consortium target BcR147A [Bacillus cereus ATCC 14579] |
| 7JRL_A | 3.81e-06 | 819 | 900 | 22 | 105 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JS4_A | 5.18e-06 | 780 | 900 | 714 | 835 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40407 | 6.20e-133 | 15 | 421 | 7 | 414 | Uncharacterized protein YbbC OS=Bacillus subtilis (strain 168) OX=224308 GN=ybbC PE=3 SV=1 |
| P56957 | 5.00e-06 | 821 | 898 | 336 | 417 | Pesticidal crystal protein Cry22Aa OS=Bacillus thuringiensis OX=1428 GN=cry22Aa PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003507 | 0.881743 | 0.114005 | 0.000331 | 0.000225 | 0.000184 |