You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001185_01068
You are here: Home > Sequence: MGYG000001185_01068
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
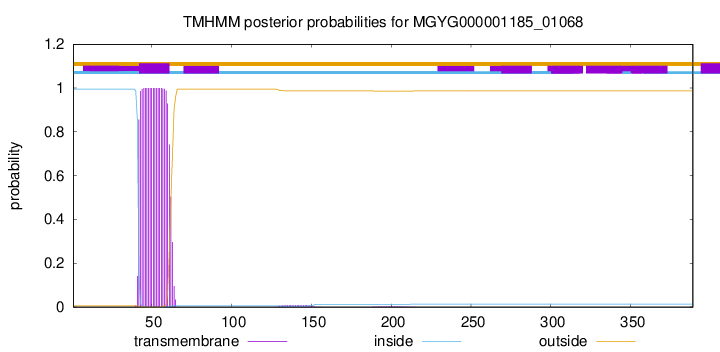
TMHMM annotations
Basic Information help
| Species | CAG-269 sp900554565 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; CAG-269; CAG-269 sp900554565 | |||||||||||
| CAZyme ID | MGYG000001185_01068 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A/1B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8665; End: 9834 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 85 | 263 | 3.5e-60 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 3.70e-75 | 75 | 385 | 53 | 344 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 3.43e-73 | 84 | 263 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG5009 | MrcA | 1.79e-59 | 76 | 344 | 47 | 298 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| PRK11636 | mrcA | 1.25e-43 | 69 | 276 | 39 | 243 | penicillin-binding protein 1a; Provisional |
| TIGR02071 | PBP_1b | 1.71e-40 | 88 | 384 | 136 | 421 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNU66566.1 | 3.17e-62 | 78 | 385 | 76 | 388 |
| QPJ85883.1 | 1.95e-59 | 73 | 381 | 55 | 382 |
| AEY67746.1 | 3.59e-59 | 72 | 385 | 71 | 389 |
| ACL77170.1 | 3.82e-59 | 72 | 385 | 71 | 389 |
| ABG87192.1 | 9.42e-59 | 73 | 381 | 67 | 390 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NB6_A | 5.43e-33 | 99 | 277 | 24 | 198 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 2OQO_A | 1.48e-32 | 99 | 277 | 24 | 198 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 5U2G_A | 2.85e-31 | 69 | 349 | 13 | 276 | 2.6Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20],5U2G_B 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20] |
| 7U4H_A | 5.48e-31 | 76 | 349 | 19 | 313 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 5ZZK_A | 4.89e-30 | 86 | 280 | 48 | 244 | ChainA, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0SRL7 | 1.88e-59 | 73 | 381 | 67 | 390 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain SM101 / Type A) OX=289380 GN=pbpA PE=3 SV=1 |
| Q8XJ01 | 2.32e-59 | 73 | 381 | 67 | 390 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=pbpA PE=3 SV=1 |
| Q0TNZ8 | 2.87e-59 | 73 | 381 | 67 | 390 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 2.86e-54 | 72 | 381 | 51 | 369 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 4.02e-54 | 72 | 381 | 51 | 369 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
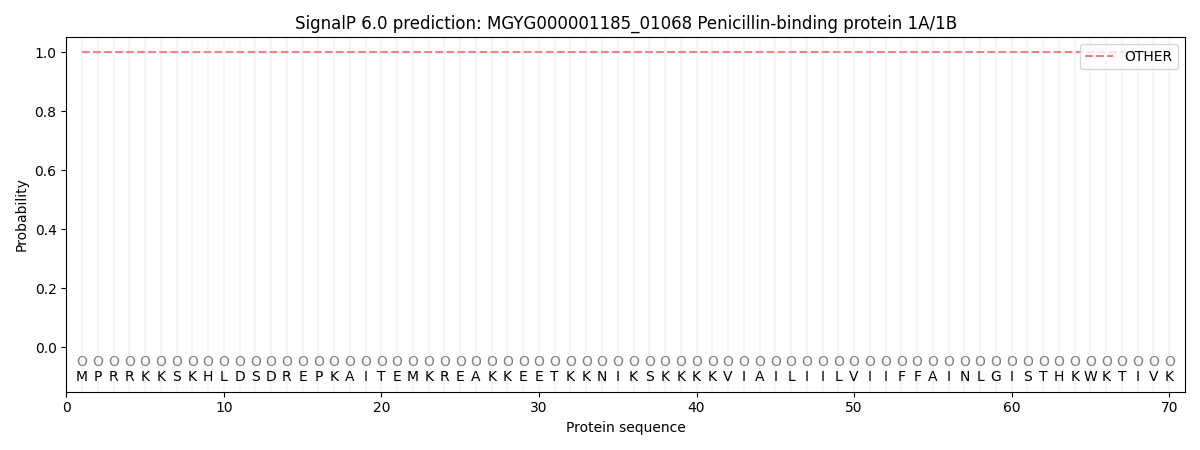
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999988 | 0.000036 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |